+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Cohesin ATPase module | ||||||||||||||||||||||||
 Map data Map data | cryo-EM map of the engaged human Cohesin ATPase module | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | 3D genome organization / Chromatin / Cohesin / ATPase activity / ATPase cycle / Cell cycle / DNA binding / DNA BINDING PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to DNA damage checkpoint signaling / negative regulation of mitotic metaphase/anaphase transition / Cohesin Loading onto Chromatin / meiotic cohesin complex / Establishment of Sister Chromatid Cohesion / establishment of meiotic sister chromatid cohesion / cohesin complex / mitotic cohesin complex / positive regulation of sister chromatid cohesion / negative regulation of glial cell apoptotic process ...response to DNA damage checkpoint signaling / negative regulation of mitotic metaphase/anaphase transition / Cohesin Loading onto Chromatin / meiotic cohesin complex / Establishment of Sister Chromatid Cohesion / establishment of meiotic sister chromatid cohesion / cohesin complex / mitotic cohesin complex / positive regulation of sister chromatid cohesion / negative regulation of glial cell apoptotic process / negative regulation of G2/M transition of mitotic cell cycle / mediator complex binding / replication-born double-strand break repair via sister chromatid exchange / establishment of mitotic sister chromatid cohesion / lateral element / chromatin looping / reciprocal meiotic recombination / microtubule motor activity / sister chromatid cohesion / mitotic sister chromatid cohesion / negative regulation of interleukin-1 beta production / stem cell population maintenance / mitotic spindle pole / lncRNA binding / dynein complex binding / regulation of DNA replication / mitotic sister chromatid segregation / positive regulation of interleukin-10 production / negative regulation of tumor necrosis factor production / somatic stem cell population maintenance / chromosome, centromeric region / mitotic spindle assembly / SUMOylation of DNA damage response and repair proteins / beta-tubulin binding / cis-regulatory region sequence-specific DNA binding / protein localization to chromatin / Meiotic synapsis / Resolution of Sister Chromatid Cohesion / condensed nuclear chromosome / meiotic cell cycle / chromosome segregation / response to radiation / kinetochore / nuclear matrix / spindle pole / Separation of Sister Chromatids / mitotic cell cycle / double-strand break repair / chromosome / double-stranded DNA binding / midbody / DNA recombination / Estrogen-dependent gene expression / DNA-binding transcription factor binding / negative regulation of neuron apoptotic process / response to hypoxia / protein heterodimerization activity / cell division / DNA repair / apoptotic process / chromatin binding / regulation of transcription by RNA polymerase II / chromatin / ATP hydrolysis activity / DNA binding / RNA binding / nucleoplasm / ATP binding / membrane / nucleus / cytosol Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||
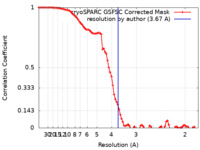
| Method | single particle reconstruction / cryo EM / Resolution: 3.67 Å | ||||||||||||||||||||||||
 Authors Authors | Landwerlin P / Durand A / Diebold-Durand M-L / Romier C | ||||||||||||||||||||||||
| Funding support |  France, 7 items France, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2024 Journal: Cell Rep / Year: 2024Title: The cohesin ATPase cycle is mediated by specific conformational dynamics and interface plasticity of SMC1A and SMC3 ATPase domains. Authors: Marina Vitoria Gomes / Pauline Landwerlin / Marie-Laure Diebold-Durand / Tajith B Shaik / Alexandre Durand / Edouard Troesch / Chantal Weber / Karl Brillet / Marianne Victoria Lemée / ...Authors: Marina Vitoria Gomes / Pauline Landwerlin / Marie-Laure Diebold-Durand / Tajith B Shaik / Alexandre Durand / Edouard Troesch / Chantal Weber / Karl Brillet / Marianne Victoria Lemée / Christophe Decroos / Ludivine Dulac / Pierre Antony / Erwan Watrin / Eric Ennifar / Christelle Golzio / Christophe Romier /  Abstract: Cohesin is key to eukaryotic genome organization and acts throughout the cell cycle in an ATP-dependent manner. The mechanisms underlying cohesin ATPase activity are poorly understood. Here, we ...Cohesin is key to eukaryotic genome organization and acts throughout the cell cycle in an ATP-dependent manner. The mechanisms underlying cohesin ATPase activity are poorly understood. Here, we characterize distinct steps of the human cohesin ATPase cycle and show that the SMC1A and SMC3 ATPase domains undergo specific but concerted structural rearrangements along this cycle. Specifically, whereas the proximal coiled coil of the SMC1A ATPase domain remains conformationally stable, that of the SMC3 displays an intrinsic flexibility. The ATP-dependent formation of the heterodimeric SMC1A/SMC3 ATPase module (engaged state) favors this flexibility, which is counteracted by NIPBL and DNA binding (clamped state). Opening of the SMC3/RAD21 interface (open-engaged state) stiffens the SMC3 proximal coiled coil, thus constricting together with that of SMC1A the ATPase module DNA-binding chamber. The plasticity of the ATP-dependent interface between the SMC1A and SMC3 ATPase domains enables these structural rearrangements while keeping the ATP gate shut. VIDEO ABSTRACT. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17331.map.gz emd_17331.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17331-v30.xml emd-17331-v30.xml emd-17331.xml emd-17331.xml | 29.5 KB 29.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17331_fsc.xml emd_17331_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_17331.png emd_17331.png | 88.8 KB | ||
| Filedesc metadata |  emd-17331.cif.gz emd-17331.cif.gz | 7.3 KB | ||
| Others |  emd_17331_half_map_1.map.gz emd_17331_half_map_1.map.gz emd_17331_half_map_2.map.gz emd_17331_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17331 http://ftp.pdbj.org/pub/emdb/structures/EMD-17331 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17331 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17331 | HTTPS FTP |
-Related structure data
| Related structure data |  8p0aMC  8pq5C  8ro6C  8ro7C  8ro8C  8ro9C  8roaC  8robC  8rocC  8rodC  8roeC  8rofC  8rogC  8rohC  8roiC  8rojC  8rokC  8rolC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17331.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17331.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM map of the engaged human Cohesin ATPase module | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.901 Å | ||||||||||||||||||||||||||||||||||||
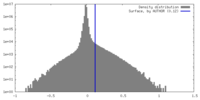
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Human Cohesin ATP-engaged ATPase module
| Entire | Name: Human Cohesin ATP-engaged ATPase module |
|---|---|
| Components |
|
-Supramolecule #1: Human Cohesin ATP-engaged ATPase module
| Supramolecule | Name: Human Cohesin ATP-engaged ATPase module / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Structural maintenance of chromosomes protein 1A
| Macromolecule | Name: Structural maintenance of chromosomes protein 1A / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.443246 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGFLKLIEIE NFKSYKGRQI IGPFQRFTAI IGPNGSGKSN LMDAISFVLG EKTSNLRVKT LRDLIHGAPV GKPAANRAFV SMVYSEEGA EDRTFARVIV GGSSEYKINN KVVQLHEYSE ELEKLGILIK ARNFLVFQGA VESIAMKNPK ERTALFEEIS R SGELAQEY ...String: MGFLKLIEIE NFKSYKGRQI IGPFQRFTAI IGPNGSGKSN LMDAISFVLG EKTSNLRVKT LRDLIHGAPV GKPAANRAFV SMVYSEEGA EDRTFARVIV GGSSEYKINN KVVQLHEYSE ELEKLGILIK ARNFLVFQGA VESIAMKNPK ERTALFEEIS R SGELAQEY DKRKKEMVKA EEDTQFNYHR KKNIAAERKE AKESSKHPTS LVPRGSDAQA EEEIKQEMNT LQQKLNEQQS VL QRIAAPN MKAMEKLESV RDKFQETSDE FEAARKRAKK AKQAFEQIKK ERFDRFNACF ESVATNIDEI YKALSRNSSA QAF LGPENP EEPYLDGINY NCVAPGKRFR PMDNLSGGEK TVAALALLFA IHSYKPAPFF VLDQIDAALD NTNIGKVANY IKEQ STCNF QAIVISLKEE FYTKAESLIG VYPEQGDCVI SKVLTFDLTK YPDANPNPNE Q UniProtKB: Structural maintenance of chromosomes protein 1A, Structural maintenance of chromosomes protein 1A |
-Macromolecule #2: Structural maintenance of chromosomes protein 3
| Macromolecule | Name: Structural maintenance of chromosomes protein 3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.391367 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MYIKQVIIQG FRSYRDQTIV DPFSSKHNVI VGRNGSGKSN FFYAIQFVLS DEFSHLRPEQ RLALLHEGTG PRVISAFVEI IFDNSDNRL PIDKEEVSLR RVIGAKKDQY FLDKKMVTKN DVMNLLESAG FSRSNPYYIV KQGKINQMAT APDSQRLKLL R EVAGTRVY ...String: MYIKQVIIQG FRSYRDQTIV DPFSSKHNVI VGRNGSGKSN FFYAIQFVLS DEFSHLRPEQ RLALLHEGTG PRVISAFVEI IFDNSDNRL PIDKEEVSLR RVIGAKKDQY FLDKKMVTKN DVMNLLESAG FSRSNPYYIV KQGKINQMAT APDSQRLKLL R EVAGTRVY DERKEESISL MKETEGKREK INELLKYIEE RLHTLEEEKE ELAGSGSLVP RGSGSYSHVN KKALDQFVNF SE QKEKLIK RQEELDRGYK SIMELMNVLE LRKYEAIQLT FKQVSKNFSE VFQKLVPGGK ATLVMKKGDV EGSQSQDEGE GSG ESERGS GSQSSVPSVD QFTGVGIRVS FTGKQGEMRE MQQLSGGQKS LVALALIFAI QKCDPAPFYL FDQIDQALDA QHRK AVSDM IMELAVHAQF ITTTFRPELL ESADKFYGVK FRNKVSHIDV ITAEMAKDFV EDDTTHG UniProtKB: Structural maintenance of chromosomes protein 3, Structural maintenance of chromosomes protein 3 |
-Macromolecule #3: 64-kDa C-terminal product
| Macromolecule | Name: 64-kDa C-terminal product / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.203648 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKRTQQMLHG LQRALAKTGA ESISLLELCR NTNRKQAAAK FYSFLVLKKQ QAIELTQEEP YSDIIATPGP RFHGSLEVLF Q UniProtKB: Double-strand-break repair protein rad21 homolog |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation #1
Sample preparation #1
| Preparation ID | 1 |
|---|---|
| Buffer | pH: 8.2 |
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Film type ID: 1 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Sample preparation #2
Sample preparation #2
| Preparation ID | 2 |
|---|---|
| Buffer | pH: 8.2 |
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Film type ID: 1 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Sample preparation #3
Sample preparation #3
| Preparation ID | 3 |
|---|---|
| Buffer | pH: 8.2 |
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Film type ID: 1 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | TFS GLACIOS |
| Image recording | Image recording ID: 1 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 4146 / Average electron dose: 44.4 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 30.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
- Electron microscopy #1~
Electron microscopy #1~
| Microscopy ID | 1 |
|---|---|
| Microscope | TFS GLACIOS |
| Image recording | Image recording ID: 2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 5574 / Average electron dose: 63.85 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 30.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
- Electron microscopy #1~~
Electron microscopy #1~~
| Microscopy ID | 1 |
|---|---|
| Microscope | TFS GLACIOS |
| Image recording | Image recording ID: 3 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 1593 / Average electron dose: 43.61 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 30.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)