+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 20S proteasome & CBR3 complex | |||||||||
 Map data Map data | Rat 20S proteasome & Human CBR3 complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / PROTEIN BINDING | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
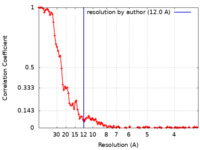
| Method | single particle reconstruction / cryo EM / Resolution: 12.0 Å | |||||||||
 Authors Authors | Deshmukh FK / Sharon M | |||||||||
| Funding support |  Israel, 1 items Israel, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Allosteric regulation of the 20S proteasome by the Catalytic Core Regulators (CCRs) family. Authors: Fanindra Kumar Deshmukh / Gili Ben-Nissan / Maya A Olshina / Maria G Füzesi-Levi / Caley Polkinghorn / Galina Arkind / Yegor Leushkin / Irit Fainer / Sarel J Fleishman / Dan Tawfik / Michal Sharon /  Abstract: Controlled degradation of proteins is necessary for ensuring their abundance and sustaining a healthy and accurately functioning proteome. One of the degradation routes involves the uncapped 20S ...Controlled degradation of proteins is necessary for ensuring their abundance and sustaining a healthy and accurately functioning proteome. One of the degradation routes involves the uncapped 20S proteasome, which cleaves proteins with a partially unfolded region, including those that are damaged or contain intrinsically disordered regions. This degradation route is tightly controlled by a recently discovered family of proteins named Catalytic Core Regulators (CCRs). Here, we show that CCRs function through an allosteric mechanism, coupling the physical binding of the PSMB4 β-subunit with attenuation of the complex's three proteolytic activities. In addition, by dissecting the structural properties that are required for CCR-like function, we could recapitulate this activity using a designed protein that is half the size of natural CCRs. These data uncover an allosteric path that does not involve the proteasome's enzymatic subunits but rather propagates through the non-catalytic subunit PSMB4. This way of 20S proteasome-specific attenuation opens avenues for decoupling the 20S and 26S proteasome degradation pathways as well as for developing selective 20S proteasome inhibitors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17118.map.gz emd_17118.map.gz | 8.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17118-v30.xml emd-17118-v30.xml emd-17118.xml emd-17118.xml | 12.8 KB 12.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17118_fsc.xml emd_17118_fsc.xml | 13 KB | Display |  FSC data file FSC data file |
| Images |  emd_17118.png emd_17118.png | 38.2 KB | ||
| Others |  emd_17118_half_map_1.map.gz emd_17118_half_map_1.map.gz emd_17118_half_map_2.map.gz emd_17118_half_map_2.map.gz | 65.6 MB 65.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17118 http://ftp.pdbj.org/pub/emdb/structures/EMD-17118 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17118 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17118 | HTTPS FTP |
-Validation report
| Summary document |  emd_17118_validation.pdf.gz emd_17118_validation.pdf.gz | 751.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17118_full_validation.pdf.gz emd_17118_full_validation.pdf.gz | 750.7 KB | Display | |
| Data in XML |  emd_17118_validation.xml.gz emd_17118_validation.xml.gz | 17 KB | Display | |
| Data in CIF |  emd_17118_validation.cif.gz emd_17118_validation.cif.gz | 22.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17118 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17118 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17118 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17118 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17118.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17118.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Rat 20S proteasome & Human CBR3 complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.7 Å | ||||||||||||||||||||||||||||||||||||
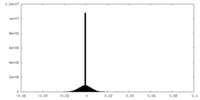
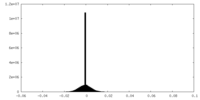
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_17118_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
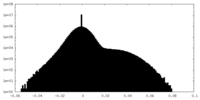
| Density Histograms |
-Half map: #2
| File | emd_17118_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
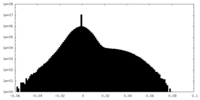
| Density Histograms |
- Sample components
Sample components
-Entire : Rat 20S proteasome complex with CBR3
| Entire | Name: Rat 20S proteasome complex with CBR3 |
|---|---|
| Components |
|
-Supramolecule #1: Rat 20S proteasome complex with CBR3
| Supramolecule | Name: Rat 20S proteasome complex with CBR3 / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: Human CBR3
| Supramolecule | Name: Human CBR3 / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Number real images: 596 / Average exposure time: 60.0 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: DIFFRACTION / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)