+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | AApoAII amyloid fibril Morphology II (ex vivo) | |||||||||
 Map data Map data | AApoAII fibril density map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | amyloid / protein fibril / systemic amyloidosis / misfolding disease / helical | |||||||||
| Function / homology | Apolipoprotein A-II (ApoA-II) / Apolipoprotein A-II (ApoA-II) superfamily / Apolipoprotein A-II (ApoA-II) / lipoprotein metabolic process / high-density lipoprotein particle / lipid transport / lipid binding / Apolipoprotein A-II Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
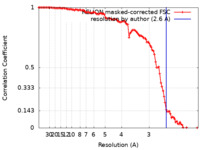
| Method | helical reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Andreotti G / Schmidt M / Faendrich M | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2024 Journal: J Mol Biol / Year: 2024Title: Insights into the Structural Basis of Amyloid Resistance Provided by Cryo-EM Structures of AApoAII Amyloid Fibrils. Authors: Giada Andreotti / Julian Baur / Marijana Ugrina / Peter Benedikt Pfeiffer / Max Hartmann / Sebastian Wiese / Hiroki Miyahara / Keiichi Higuchi / Nadine Schwierz / Matthias Schmidt / Marcus Fändrich /   Abstract: Amyloid resistance is the inability or the reduced susceptibility of an organism to develop amyloidosis. In this study we have analysed the molecular basis of the resistance to systemic AApoAII ...Amyloid resistance is the inability or the reduced susceptibility of an organism to develop amyloidosis. In this study we have analysed the molecular basis of the resistance to systemic AApoAII amyloidosis, which arises from the formation of amyloid fibrils from apolipoprotein A-II (ApoA-II). The disease affects humans and animals, including SAMR1C mice that express the C allele of ApoA-II protein, whereas other mouse strains are resistant to development of amyloidosis due to the expression of other ApoA-II alleles, such as ApoA-IIF. Using cryo-electron microscopy, molecular dynamics simulations and other methods, we have determined the structures of pathogenic AApoAII amyloid fibrils from SAMR1C mice and analysed the structural effects of ApoA-IIF-specific mutational changes. Our data show that these changes render ApoA-IIF incompatible with the specific fibril morphologies, with which ApoA-II protein can become pathogenic in vivo. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17104.map.gz emd_17104.map.gz | 834 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17104-v30.xml emd-17104-v30.xml emd-17104.xml emd-17104.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17104_fsc.xml emd_17104_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_17104.png emd_17104.png | 60.5 KB | ||
| Masks |  emd_17104_msk_1.map emd_17104_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17104.cif.gz emd-17104.cif.gz | 5.6 KB | ||
| Others |  emd_17104_additional_1.map.gz emd_17104_additional_1.map.gz emd_17104_half_map_1.map.gz emd_17104_half_map_1.map.gz emd_17104_half_map_2.map.gz emd_17104_half_map_2.map.gz | 5.9 MB 80.3 MB 80.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17104 http://ftp.pdbj.org/pub/emdb/structures/EMD-17104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17104 | HTTPS FTP |
-Validation report
| Summary document |  emd_17104_validation.pdf.gz emd_17104_validation.pdf.gz | 783.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17104_full_validation.pdf.gz emd_17104_full_validation.pdf.gz | 782.9 KB | Display | |
| Data in XML |  emd_17104_validation.xml.gz emd_17104_validation.xml.gz | 17.8 KB | Display | |
| Data in CIF |  emd_17104_validation.cif.gz emd_17104_validation.cif.gz | 23.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17104 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17104 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17104 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17104 | HTTPS FTP |
-Related structure data
| Related structure data |  8oq4MC  8oq5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17104.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17104.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AApoAII fibril density map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
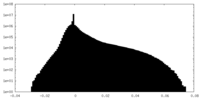
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17104_msk_1.map emd_17104_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
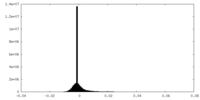
| Density Histograms |
-Additional map: Extended AApoAII fibril density map
| File | emd_17104_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Extended AApoAII fibril density map | ||||||||||||
| Projections & Slices |
| ||||||||||||
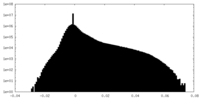
| Density Histograms |
-Half map: half map 1
| File | emd_17104_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_17104_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : AApoAII amyloid fibril
| Entire | Name: AApoAII amyloid fibril |
|---|---|
| Components |
|
-Supramolecule #1: AApoAII amyloid fibril
| Supramolecule | Name: AApoAII amyloid fibril / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: AApoAII amyloid fibrils extracted from SAMR1C mice. |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Apolipoprotein A-II
| Macromolecule | Name: Apolipoprotein A-II / type: protein_or_peptide / ID: 1 / Number of copies: 24 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.746728 KDa |
| Sequence | String: QADGQDMQSL FTQYFQSMTE YGKDLVEKAK TSEIQSQAKA YFEKTHEQLT PLVRSAGTSL VNFFSSLMNL EEKPAPAA UniProtKB: Apolipoprotein A-II |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Details: Water |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 294.15 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average exposure time: 12.0 sec. / Average electron dose: 42.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: SPOT SCAN / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8oq4: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)