+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | EcoR124I bound to dsDNA | |||||||||
 Map data Map data | Type IC restriction modification system EcoR124I bound to 38-mer dsDNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Pentameric complex / restriction-modification enzyme / restriction endonuclease / methyltransferase / ANTIVIRAL PROTEIN | |||||||||
| Biological species |  | |||||||||
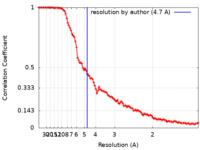
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Pradeep HN | |||||||||
| Funding support |  Czech Republic, 1 items Czech Republic, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: EcoR124I bound to dsDNA Authors: Pradeep HN | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17088.map.gz emd_17088.map.gz | 50 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17088-v30.xml emd-17088-v30.xml emd-17088.xml emd-17088.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17088_fsc.xml emd_17088_fsc.xml | 13.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_17088.png emd_17088.png | 87 KB | ||
| Others |  emd_17088_half_map_1.map.gz emd_17088_half_map_1.map.gz emd_17088_half_map_2.map.gz emd_17088_half_map_2.map.gz | 91.6 MB 91.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17088 http://ftp.pdbj.org/pub/emdb/structures/EMD-17088 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17088 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17088 | HTTPS FTP |
-Validation report
| Summary document |  emd_17088_validation.pdf.gz emd_17088_validation.pdf.gz | 865.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17088_full_validation.pdf.gz emd_17088_full_validation.pdf.gz | 865.4 KB | Display | |
| Data in XML |  emd_17088_validation.xml.gz emd_17088_validation.xml.gz | 18 KB | Display | |
| Data in CIF |  emd_17088_validation.cif.gz emd_17088_validation.cif.gz | 23.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17088 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17088 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17088 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17088 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17088.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17088.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Type IC restriction modification system EcoR124I bound to 38-mer dsDNA | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.714 Å | ||||||||||||||||||||||||||||||||||||
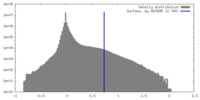
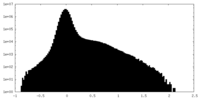
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Type IC restriction modification system EcoR124I bound to...
| File | emd_17088_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Type IC restriction modification system EcoR124I bound to 38-mer dsDNA - halfmap A | ||||||||||||
| Projections & Slices |
| ||||||||||||
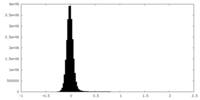
| Density Histograms |
-Half map: Type IC restriction modification system EcoR124I bound to...
| File | emd_17088_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Type IC restriction modification system EcoR124I bound to 38-mer dsDNA - halfmap B | ||||||||||||
| Projections & Slices |
| ||||||||||||
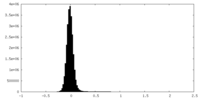
| Density Histograms |
- Sample components
Sample components
-Entire : EcoR124I+DNA
| Entire | Name: EcoR124I+DNA |
|---|---|
| Components |
|
-Supramolecule #1: EcoR124I+DNA
| Supramolecule | Name: EcoR124I+DNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 0.4 kDa/nm |
-Macromolecule #1: Type I C restriction-modification complex EcoR124I
| Macromolecule | Name: Type I C restriction-modification complex EcoR124I / type: other / ID: 1 Details: Subunit HsdR (2x); Subunit HsdM (2x); Subunit HsdS (1x) Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MTHQTHTIAE SNNFIVLDKY IKAEPTGDSY QSESDLEREL IQDLRNQGYE FISVKSQSAM LANVREQLQ NLNGVVFNDS EWRRFTEQYL DNPSDGILDK TRKIHIDYIC DFIFDDERLE N IYLIDKKN LMRNKVQIIQ QFEQAGSHAN RYDVTILVNG LPLVQIELKK ...String: MTHQTHTIAE SNNFIVLDKY IKAEPTGDSY QSESDLEREL IQDLRNQGYE FISVKSQSAM LANVREQLQ NLNGVVFNDS EWRRFTEQYL DNPSDGILDK TRKIHIDYIC DFIFDDERLE N IYLIDKKN LMRNKVQIIQ QFEQAGSHAN RYDVTILVNG LPLVQIELKK RGVAIREAFN QI HRYSKES FNSENSLFKY LQLFVISNGT DTRYFANTTK RDKNSFDFTM NWAKSDNTLI KDL KDFTAT CFQKHTLLNV LVNYSVFDSS QTLLVMRPYQ IAATERILWK IKSSFTAKNW SKPE SGGYI WHTTGSGKTL TSFKAARLAT ELDFIDKVFF VVDRKDLDYQ TMKEYQRFSP DSVNG SENT AGLKRNLDKD DNKIIVTTIQ KLNNLMKAES DLPVYNQQVV FIFDECHRSQ FGEAQK NLK KKFKRYYQFG FTGTPIFPEN ALGSETTASV FGRELHSYVI TDAIRDEKVL KFKVDYN DV RPQFKSLETE TDEKKLSAAE NQQAFLHPMR IQEITQYILN NFRQKTHRTF PGSKGFNA M LAVSSVDAAK AYYATFKRLQ EEAANKSATY KPLRIATIFS FAANEEQNAI GEISDETFD TSAMDSSAKE FLDAAIREYN SHFKTNFSTD SNGFQNYYRD LAQRVKNQDI DLLIVVGMFL TGFDAPTLN TLFVDKNLRY HGLMQAFSRT NRIYDATKTF GNIVTFRDLE RSTIDAITLF G DKNTKNVV LEKSYTEYME GFTDAATGEA KRGFMTVVSE LEQRFPDPTS IESEKEKKDF VK LFGEYLR AENILQNYDE FATLKALQQI DLSDPVAVEK FKAEHYVDDE KFAELQTIRL PAD RKIQDY RSAYNDIRDW QRREKEAEKK EKSTTDWDDV VFEVDLLKSQ EINLDYILGL IFEH NRQNK GKGEMIEEVK RLIRSSLGNR AKEGLVVDFI QQTNLDDLPD KASIIDAFFT FAQRE QQRE AEALIKEENL NEDAAKRYIR TSLKREYATE NGTELNETLP KLSPLNPQYK TKKQAV FRK SSRLLRSLKA MKMTSIQQRA ELHRQIWQIA NDVRGSVDGW DFKQYVLGAL FYRFISE NF SSYIEAGDDS ICYAKLDDSV ITDDIKDDAI KTKGYFIYPS QLFCNVAAKA NTNDRLNA D LNSIFVAIES SAYGYPSEAD IKGLFADFDT TSNRLGNTVK DKNARLAAVL KGVEGLKLG DFNEHQIDLF GDAYEFLISN YAANAGKSGG EFFTPQHVSK LIAQLAMHGQ THVNKIYDPA AGSGSLLLQ AKKQFDNHII EEGFFGQEIN HTTYNLARMN MFLHNINYDK FDIKLGNTLT E PHFRDEKP FDAIVSNPPY SVKWIGSDDP TLINDERFAP AGVLAPKSKA DFAFVLHALN YL SAKGRAA IVCFPGIFYR GGAEQKIRQY LVDNNYVETV ISLAPNLFFG TTIAVNILVL SKH KTDTNV QFIDASELFK KETNNNILTD AHIEQIMQVF ASKEDVAHLA KSVAFETVVA NDYN LSVSS YVEAKDNREI IDIAELNAEL KTTVSKIDQL RKDIDAIVAE IEGCEVQKMS EMSYL EKLL DGVEVEWLPL GEITKYEQPT KYLVKAKDYH DTYTIPVLTA GKTFILGYTN ETHGIY QAS KAPVIIFDDF TTANKWVDFD FKAKSSAMKM VTSCDDNKTL LKYVYYWLNT LPSEFAE GD HKRQWISNYS QKKIPIPCPD NPEKSLAIQS EIVRILDKFT ALTAELTAEL NMRKKQYN Y YRDQLLSFKE GEVEWKTLGE IGKWYGGGTP SKNKIEFWEN GSIPWISPKD MGRTLVDSS EDYITEEAVL HSSTKLIPAN SIAIVVRSSI LDKVLPSALI KVPATLNQDM KAVIPHENIL VKYIYHMIG SRGSDILRAA KKTGGSVASI DSKKLFSFKI PVPNINEQQR IVEILDKFDT L TNSITEGL PREIELRQKQ YEYYRDLLFS FPKPETVSNM KMTSIQQRAE LHRQIWQIAN DV RGSVDGW DFKQYVLGAL FYRFISENFS SYIEAGDDSI CYAKLDDSVI TDDIKDDAIK TKG YFIYPS QLFCNVAAKA NTNDRLNADL NSIFVAIESS AYGYPSEADI KGLFADFDTT SNRL GNTVK DKNARLAAVL KGVEGLKLGD FNEHQIDLFG DAYEFLISNY AANAGKSGGE FFTPQ HVSK LIAQLAMHGQ THVNKIYDPA AGSGSLLLQA KKQFDNHIIE EGFFGQEINH TTYNLA RMN MFLHNINYDK FDIKLGNTLT EPHFRDEKPF DAIVSNPPYS VKWIGSDDPT LINDERF AP AGVLAPKSKA DFAFVLHALN YLSAKGRAAI VCFPGIFYRG GAEQKIRQYL VDNNYVET V ISLAPNLFFG TTIAVNILVL SKHKTDTNVQ FIDASELFKK ETNNNILTDA HIEQIMQVF ASKEDVAHLA KSVAFETVVA NDYNLSVSSY VEAKDNREII DIAELNAELK TTVSKIDQLR KDIDAIVAE IEGCEVQKMT HQTHTIAESN NFIVLDKYIK AEPTGDSYQS ESDLERELIQ D LRNQGYEF ISVKSQSAML ANVREQLQNL NGVVFNDSEW RRFTEQYLDN PSDGILDKTR KI HIDYICD FIFDDERLEN IYLIDKKNLM RNKVQIIQQF EQAGSHANRY DVTILVNGLP LVQ IELKKR GVAIREAFNQ IHRYSKESFN SENSLFKYLQ LFVISNGTDT RYFANTTKRD KNSF DFTMN WAKSDNTLIK DLKDFTATCF QKHTLLNVLV NYSVFDSSQT LLVMRPYQIA ATERI LWKI KSSFTAKNWS KPESGGYIWH TTGSGKTLTS FKAARLATEL DFIDKVFFVV DRKDLD YQT MKEYQRFSPD SVNGSENTAG LKRNLDKDDN KIIVTTIQKL NNLMKAESDL PVYNQQV VF IFDECHRSQF GEAQKNLKKK FKRYYQFGFT GTPIFPENAL GSETTASVFG RELHSYVI T DAIRDEKVLK FKVDYNDVRP QFKSLETETD EKKLSAAENQ QAFLHPMRIQ EITQYILNN FRQKTHRTFP GSKGFNAMLA VSSVDAAKAY YATFKRLQEE AANKSATYKP LRIATIFSFA ANEEQNAIG EISDETFDTS AMDSSAKEFL DAAIREYNSH FKTNFSTDSN GFQNYYRDLA Q RVKNQDID LLIVVGMFLT GFDAPTLNTL FVDKNLRYHG LMQAFSRTNR IYDATKTFGN IV TFRDLER STIDAITLFG DKNTKNVVLE KSYTEYMEGF TDAATGEAKR GFMTVVSELE QRF PDPTSI ESEKEKKDFV KLFGEYLRAE NILQNYDEFA TLKALQQIDL SDPVAVEKFK AEHY VDDEK FAELQTIRLP ADRKIQDYRS AYNDIRDWQR REKEAEKKEK STTDWDDVVF EVDLL KSQE INLDYILGLI FEHNRQNKGK GEMIEEVKRL IRSSLGNRAK EGLVVDFIQQ TNLDDL PDK ASIIDAFFTF AQREQQREAE ALIKEENLNE DAAKRYIRTS LKREYATENG TELNETL PK LSPLNPQYKT KKQAVFRKSS RLLRSLKA |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20 mM NaCl, 50 mM HEPEs |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % |
| Details | This sample is heterogeneous |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)