[English] 日本語
 Yorodumi
Yorodumi- EMDB-17032: CryoEM Structure INO80 hexasome complex Arp8 module bound to DNA -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM Structure INO80 hexasome complex Arp8 module bound to DNA | |||||||||||||||
 Map data Map data | ctINO80 hexasome complex Arp8 Module unsharpened map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | ATP-dependent chromatin remodeler / DNA BINDING PROTEIN | |||||||||||||||
| Biological species |  Thermochaetoides thermophila (fungus) Thermochaetoides thermophila (fungus) | |||||||||||||||
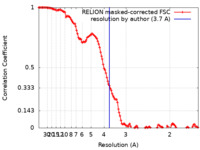
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||||||||
 Authors Authors | Zhang M / Jungblut A / Hoffmann T / Eustermann S | |||||||||||||||
| Funding support |  Germany, European Union, 4 items Germany, European Union, 4 items
| |||||||||||||||
 Citation Citation | Journal: Acta Crystallogr D Biol Crystallogr / Year: 2010 Title: Features and development of Coot. Authors: P Emsley / B Lohkamp / W G Scott / K Cowtan /  Abstract: Coot is a molecular-graphics application for model building and validation of biological macromolecules. The program displays electron-density maps and atomic models and allows model manipulations ...Coot is a molecular-graphics application for model building and validation of biological macromolecules. The program displays electron-density maps and atomic models and allows model manipulations such as idealization, real-space refinement, manual rotation/translation, rigid-body fitting, ligand search, solvation, mutations, rotamers and Ramachandran idealization. Furthermore, tools are provided for model validation as well as interfaces to external programs for refinement, validation and graphics. The software is designed to be easy to learn for novice users, which is achieved by ensuring that tools for common tasks are 'discoverable' through familiar user-interface elements (menus and toolbars) or by intuitive behaviour (mouse controls). Recent developments have focused on providing tools for expert users, with customisable key bindings, extensions and an extensive scripting interface. The software is under rapid development, but has already achieved very widespread use within the crystallographic community. The current state of the software is presented, with a description of the facilities available and of some of the underlying methods employed. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17032.map.gz emd_17032.map.gz | 40.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17032-v30.xml emd-17032-v30.xml emd-17032.xml emd-17032.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17032_fsc.xml emd_17032_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_17032.png emd_17032.png | 35.7 KB | ||
| Others |  emd_17032_half_map_1.map.gz emd_17032_half_map_1.map.gz emd_17032_half_map_2.map.gz emd_17032_half_map_2.map.gz | 40.8 MB 40.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17032 http://ftp.pdbj.org/pub/emdb/structures/EMD-17032 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17032 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17032 | HTTPS FTP |
-Validation report
| Summary document |  emd_17032_validation.pdf.gz emd_17032_validation.pdf.gz | 848.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17032_full_validation.pdf.gz emd_17032_full_validation.pdf.gz | 848 KB | Display | |
| Data in XML |  emd_17032_validation.xml.gz emd_17032_validation.xml.gz | 17.4 KB | Display | |
| Data in CIF |  emd_17032_validation.cif.gz emd_17032_validation.cif.gz | 23.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17032 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17032 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17032 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17032 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17032.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17032.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ctINO80 hexasome complex Arp8 Module unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||
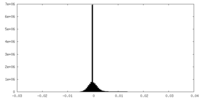
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: ctINO80 hexasome complex Arp8 Module half map
| File | emd_17032_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ctINO80 hexasome complex Arp8 Module half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
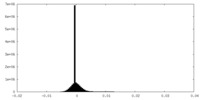
| Density Histograms |
-Half map: ctINO80 hexasome complex Arp8 Module half map
| File | emd_17032_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ctINO80 hexasome complex Arp8 Module half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
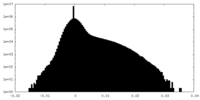
| Density Histograms |
- Sample components
Sample components
-Entire : INO80 core module in complex with hexasome
| Entire | Name: INO80 core module in complex with hexasome |
|---|---|
| Components |
|
-Supramolecule #1: INO80 core module in complex with hexasome
| Supramolecule | Name: INO80 core module in complex with hexasome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#12 Details: 11-subunit ct INO80 contains two modules (core and Arp8 module) Each module was picked and analyzed separately The core module + hexasome has an overall weight of 0.861MDa The 11-subunit ct ...Details: 11-subunit ct INO80 contains two modules (core and Arp8 module) Each module was picked and analyzed separately The core module + hexasome has an overall weight of 0.861MDa The 11-subunit ct INO80 + hexasome has an overall weight of 1.1MDa Ino80, Ies2, Ies6, Ies4,Arp6, Rvb1, Rvb2, Arp8, Arp4, Actin, Taf14 Hexasome DNA, 2xH3, 2xH4, H2A, H2B |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila (fungus) Thermochaetoides thermophila (fungus) |
| Molecular weight | Theoretical: 861 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.88 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 30mM HEPES, pH7.5 50mM NaCl 0.25mM CaCl2 0.25mM DTT 2mM ADP 3.3mM MgCl2 10mM NaF 2mM AlCl3 0.05% octyl-beta-glucoside |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 15 sec. / Details: 10% Oxygene 90% Argon |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV Details: wait time of 5s, blot force at 3, and a blot time of 2s with Whatman blotting paper (Cytiva, CAT No. 10311807). |
| Details | 11-subunit ctINO80 reconstituted with hexasome |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 15384 / Average electron dose: 50.36 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)