[English] 日本語
 Yorodumi
Yorodumi- EMDB-16871: Subtomogram average of short bridges of the yeast ER-mitochondria... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average of short bridges of the yeast ER-mitochondria encounter structure (ERMES). The population half containing shorter bridge structures was averaged. | ||||||||||||||||||||||||
 Map data Map data | Subtomogram average (STA) map of short ERMES bridge structures. The data set of all bridges was split into halves according to bridge length. The map has been aligned to the STA map of all bridges. | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | ERMES / lipid transfer protein / membrane contact sites / SMP domains / LIPID TRANSPORT | ||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||
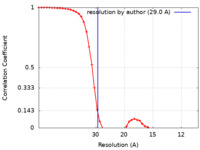
| Method | subtomogram averaging / cryo EM / Resolution: 29.0 Å | ||||||||||||||||||||||||
 Authors Authors | Wozny MR / Di Luca A / Morado DR / Picco A / Khaddaj R / Campomanes P / Ivanovic L / Hoffmann PC / Miller EA / Vanni S / Kukulski W | ||||||||||||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  Canada, Canada,  Switzerland, European Union, 7 items Switzerland, European Union, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: In situ architecture of the ER-mitochondria encounter structure. Authors: Michael R Wozny / Andrea Di Luca / Dustin R Morado / Andrea Picco / Rasha Khaddaj / Pablo Campomanes / Lazar Ivanović / Patrick C Hoffmann / Elizabeth A Miller / Stefano Vanni / Wanda Kukulski /      Abstract: The endoplasmic reticulum and mitochondria are main hubs of eukaryotic membrane biogenesis that rely on lipid exchange via membrane contact sites, but the underpinning mechanisms remain poorly ...The endoplasmic reticulum and mitochondria are main hubs of eukaryotic membrane biogenesis that rely on lipid exchange via membrane contact sites, but the underpinning mechanisms remain poorly understood. In yeast, tethering and lipid transfer between the two organelles is mediated by the endoplasmic reticulum-mitochondria encounter structure (ERMES), a four-subunit complex of unresolved stoichiometry and architecture. Here we determined the molecular organization of ERMES within Saccharomyces cerevisiae cells using integrative structural biology by combining quantitative live imaging, cryo-correlative microscopy, subtomogram averaging and molecular modelling. We found that ERMES assembles into approximately 25 discrete bridge-like complexes distributed irregularly across a contact site. Each bridge consists of three synaptotagmin-like mitochondrial lipid binding protein domains oriented in a zig-zag arrangement. Our molecular model of ERMES reveals a pathway for lipids. These findings resolve the in situ supramolecular architecture of a major inter-organelle lipid transfer machinery and provide a basis for the mechanistic understanding of lipid fluxes in eukaryotic cells. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16871.map.gz emd_16871.map.gz | 6.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16871-v30.xml emd-16871-v30.xml emd-16871.xml emd-16871.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16871_fsc.xml emd_16871_fsc.xml | 5.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_16871.png emd_16871.png | 191 KB | ||
| Others |  emd_16871_additional_1.map.gz emd_16871_additional_1.map.gz emd_16871_additional_2.map.gz emd_16871_additional_2.map.gz emd_16871_additional_3.map.gz emd_16871_additional_3.map.gz emd_16871_half_map_1.map.gz emd_16871_half_map_1.map.gz emd_16871_half_map_2.map.gz emd_16871_half_map_2.map.gz | 390 KB 388.8 KB 6.1 MB 6.1 MB 6.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16871 http://ftp.pdbj.org/pub/emdb/structures/EMD-16871 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16871 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16871 | HTTPS FTP |
-Validation report
| Summary document |  emd_16871_validation.pdf.gz emd_16871_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16871_full_validation.pdf.gz emd_16871_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_16871_validation.xml.gz emd_16871_validation.xml.gz | 9.8 KB | Display | |
| Data in CIF |  emd_16871_validation.cif.gz emd_16871_validation.cif.gz | 13.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16871 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16871 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16871 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16871 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16871.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16871.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average (STA) map of short ERMES bridge structures. The data set of all bridges was split into halves according to bridge length. The map has been aligned to the STA map of all bridges. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.368 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Half map b, masked. Together with masked half...
| File | emd_16871_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map b, masked. Together with masked half map a, this was used for resolution estimate. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Half map a, masked. Together with masked half...
| File | emd_16871_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map a, masked. Together with masked half map b, this was used for resolution estimate. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: This is the main STA map of short...
| File | emd_16871_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the main STA map of short bridges, not aligned to the STA map of all bridges. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map a, unmasked
| File | emd_16871_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map a, unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map b, unmasked
| File | emd_16871_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map b, unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Budding yeast cells expressing Mdm34-mNeonGreen, a component of t...
| Entire | Name: Budding yeast cells expressing Mdm34-mNeonGreen, a component of the ER-mitochondria encounter structure (ERMES). |
|---|---|
| Components |
|
-Supramolecule #1: Budding yeast cells expressing Mdm34-mNeonGreen, a component of t...
| Supramolecule | Name: Budding yeast cells expressing Mdm34-mNeonGreen, a component of the ER-mitochondria encounter structure (ERMES). type: cell / ID: 1 / Parent: 0 Details: Yeast cells were cryo-FIB-milled, imaged by cryo-fluorescence microscopy and electron cryo-tomograms were acquired at localisations of Mdm34-mNeonGreen fluorescent signals. |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 6 Details: Synthetic complete medium without tryptophan, with 2% glucose and 15% dextran |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
| Details | Plunge-frozen grids with yeast cells were subjected to cryo-FIB milling followed by cryo-fluorescence microscopy prior to cryo-ET. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 6.0 µm / Nominal defocus min: 3.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)