+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the SNV L protein bound to 5' RNA | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Negative-strand viruses / Bunyaviruses / Hantaviruses / RNA-dependent RNA polymerase / viral genome replication and transcription / VIRUS PROTEIN / VIRAL PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcap snatching / endonuclease activity / host cell perinuclear region of cytoplasm / RNA-directed RNA polymerase / viral RNA genome replication / RNA-dependent RNA polymerase activity / DNA-templated transcription Similarity search - Function | ||||||||||||||||||
| Biological species |  Sin Nombre orthohantavirus Sin Nombre orthohantavirus | ||||||||||||||||||
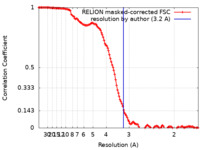
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||||||||
 Authors Authors | Meier K / Thorkelsson SR / Durieux Trouilleton Q / Vogel D / Yu D / Kosinski J / Cusack S / Malet H / Grunewald K / Quemin ERJ / Rosenthal M | ||||||||||||||||||
| Funding support |  Germany, Germany,  France, 5 items France, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2023 Journal: PLoS Pathog / Year: 2023Title: Structural and functional characterization of the Sin Nombre virus L protein. Authors: Kristina Meier / Sigurdur R Thorkelsson / Quentin Durieux Trouilleton / Dominik Vogel / Dingquan Yu / Jan Kosinski / Stephen Cusack / Hélène Malet / Kay Grünewald / Emmanuelle R J Quemin / Maria Rosenthal /   Abstract: The Bunyavirales order is a large and diverse group of segmented negative-strand RNA viruses. Several virus families within this order contain important human pathogens, including Sin Nombre virus ...The Bunyavirales order is a large and diverse group of segmented negative-strand RNA viruses. Several virus families within this order contain important human pathogens, including Sin Nombre virus (SNV) of the Hantaviridae. Despite the high epidemic potential of bunyaviruses, specific medical countermeasures such as vaccines or antivirals are missing. The multifunctional ~250 kDa L protein of hantaviruses, amongst other functional domains, harbors the RNA-dependent RNA polymerase (RdRp) and an endonuclease and catalyzes transcription as well as replication of the viral RNA genome, making it a promising therapeutic target. The development of inhibitors targeting these key processes requires a profound understanding of the catalytic mechanisms. Here, we established expression and purification protocols of the full-length SNV L protein bearing the endonuclease mutation K124A. We applied different biochemical in vitro assays to provide an extensive characterization of the different enzymatic functions as well as the capacity of the hantavirus L protein to interact with the viral RNA. By using single-particle cryo-EM, we obtained a 3D model including the L protein core region containing the RdRp, in complex with the 5' promoter RNA. This first high-resolution model of a New World hantavirus L protein shows striking similarity to related bunyavirus L proteins. The interaction of the L protein with the 5' RNA observed in the structural model confirms our hypothesis of protein-RNA binding based on our biochemical data. Taken together, this study provides an excellent basis for future structural and functional studies on the hantavirus L protein and for the development of antiviral compounds. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16670.map.gz emd_16670.map.gz | 11.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16670-v30.xml emd-16670-v30.xml emd-16670.xml emd-16670.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16670_fsc.xml emd_16670_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_16670.png emd_16670.png | 78.1 KB | ||
| Others |  emd_16670_half_map_1.map.gz emd_16670_half_map_1.map.gz emd_16670_half_map_2.map.gz emd_16670_half_map_2.map.gz | 140.7 MB 140.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16670 http://ftp.pdbj.org/pub/emdb/structures/EMD-16670 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16670 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16670 | HTTPS FTP |
-Validation report
| Summary document |  emd_16670_validation.pdf.gz emd_16670_validation.pdf.gz | 769.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16670_full_validation.pdf.gz emd_16670_full_validation.pdf.gz | 768.6 KB | Display | |
| Data in XML |  emd_16670_validation.xml.gz emd_16670_validation.xml.gz | 20.1 KB | Display | |
| Data in CIF |  emd_16670_validation.cif.gz emd_16670_validation.cif.gz | 26.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16670 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16670 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16670 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16670 | HTTPS FTP |
-Related structure data
| Related structure data |  8ci5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16670.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16670.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_16670_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
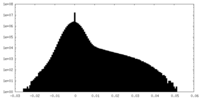
| Projections & Slices |
| ||||||||||||
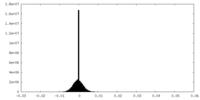
| Density Histograms |
-Half map: #1
| File | emd_16670_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
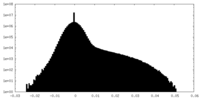
| Projections & Slices |
| ||||||||||||
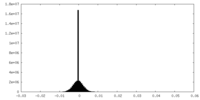
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of Sin nombre virus L protein bound to 5' RNA
| Entire | Name: Structure of Sin nombre virus L protein bound to 5' RNA |
|---|---|
| Components |
|
-Supramolecule #1: Structure of Sin nombre virus L protein bound to 5' RNA
| Supramolecule | Name: Structure of Sin nombre virus L protein bound to 5' RNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Sin Nombre orthohantavirus Sin Nombre orthohantavirus |
| Molecular weight | Theoretical: 254 KDa |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Sin Nombre orthohantavirus Sin Nombre orthohantavirus |
| Molecular weight | Theoretical: 246.846797 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MEKYREIHQR VKEIPPGGAS ALECLDLLDR LYAVRHDVVD QMIKHDWSDN KDMERPIGQV LLMAGVPNDI IQGMEKKVIP TSPSGQILK SFFRMTPDNY KITGALIEFI EVTVTADVAK GIREAKLKYE SGLQFIESLL SQEHKKGNIN QVYKITFDVV A VKTDGSNI ...String: MEKYREIHQR VKEIPPGGAS ALECLDLLDR LYAVRHDVVD QMIKHDWSDN KDMERPIGQV LLMAGVPNDI IQGMEKKVIP TSPSGQILK SFFRMTPDNY KITGALIEFI EVTVTADVAK GIREAKLKYE SGLQFIESLL SQEHKKGNIN QVYKITFDVV A VKTDGSNI STQWPSRRND GVVQHMRLVQ ADINYVREHL IKPDERASLE AMFNLKFHVG GPKLRYFNIP DYKPQSLCQP EI TNLIQYC KHWLTEDHDF VFKEVTGNNV MSSFENNEDV YMSRYKESRK PRNFLLIQGS IQGPYLPSTI SSDQCDTRIG CLE VLKVHP ETPVQAIAVD MAYKYMELNR DEIINYYNPR VHFQATQSVK EPGTFKLGLS QLNPMSKSIL DQVGKHKSEK GLFG EPLES INISSQIQQN ECSRIIESIL SNLEINVGEV TMNLANPRKT TGVDELLGKF YENELSKYLI SILRKTAAWH IGHLI RDIT ESLIAHAGLK RSKYWSIHAY DHGGVILFIL PSKSLEVVGS YIRYFTVFKD GIGLIDEENL DSKVDIDGVQ WCFSKV MSI DLNRLLALNI AFEKALLATA TWFQYYTEDQ GHFPLQHALR SVFSFHFLLC VSQKMKICAI FDNLRYLIPA VTSLYSG YE LLIEKFFERP FKSALEVYLY NIIKALLISL AQNNKVRFYS KVRLLGLTVD HSTVGASGVY PSLMSRVVYK HYRSLISE A TTCFFLFEKG LHGNLNEEAK IHLETVEWAR KFEAKERKYG DILMREGYTI DAIRVGDVQV EQQLFCQEVV ELSAEELNK YLQAKSQVLS SNIMNKHWDK PYFSQTRNIS LKGMSGALQE DGHLAASVTL IEAIRFLNRS QTNPNVIDMY EQTKQHKAQA RIVRKYQRT EADRGFFITT LPTRVRLEII EDYYDAIARV VPEEYISYGG DKKILNIQTA LEKALRWASG SSEITTSTGN V IKFKRRLM YVSADATKWS PGDNSAKFKR FTQALYDGLS DEKLKCCVVD ALRHVYETEF FMSRKLHRYI DSMDEHSEAV QD FLDFFKG GVSATVKGNW LQGNLNKCSS LFGAAVSLLF RRIWAELFPE LECFFEFAHH SDDALFIYGY LEPEDDGTDW FLY VSQQIQ AGNYHWHAVN QEMWKSMFNL HEHLLLMGSI KVSPKKTTVS PTNAEFLSTF FEGCAVSIPF IKILLGSLSD LPGL GFFDD LAAAQSRCVK AMDLGASPQL AQLAVVICTS KVERLYGTAD GMVNSPVAFL KVTKAHVPIP LGGDGSMSIM ELATA GIGM ADKNILKQAF YSYKHTRRDG DRYVLGLFKF LMSLSEDVFQ HDRLGEFSFV GKVQWKVFTP KNEFEFYDQF SQSYLK SWT NQHPVYDYII PRGRDNLLVY LVRKLNDPSI VTAMTMQSPL QLRFRMQAKQ HMKVCKLDGE WVTFREVLAA ADSFATK YN PTEKDLDLFN TLVSCTFSKE YAWKDFLNEV RCEVVPTKHV HRSKIARTFT VREKDQAIQN PITAVIGYKY ASTVDEIS D VLDSSFFPDS LSADLQVMKE GVYRELGLDI GLPEVLKRIA PLLYKAGRSR VVIVEGNVEG TAESICSYWL RSMSLVKTI KVRPKKEVLR AVSLYSTKEN IGLQDDVAAT RLCIEVWRWC KANDQNVNDW LNALYFEKQT LMDWVERFRR KGVVPVDPEI QCIALLLYD VLGYKSVLQM QANRRAYSGK QYDAYCVQTY NEETKLYEGD LRVTFNFGLD CARLEIFWDK KEYILETSIT Q RHVLKLMM EEVTQELLRC GMRFKTEQVS HTKSLVLFKT ESGFEWGKPN VPCIVFKHCA LRTGLRTKQA INKEFMINVQ AD GFRAIAQ MDVESPRFLL AHAYHTLRDV RYQAVQAVGN VWFQTNQHKL FINPIISSGL LENFMKGLPA AIPPAAYSLI MNK AKISVD LFMFNELLAL VNPRNVLNLD GIEETSEGYS TVTSISSRQW SEEVSLMADD NIDDEEEFTI ALDDIDFEQI NLDE DIQHF LQDESAYTGD LTIQTEEVEV KRIRGVTRVL EPVKLIKSWV SKGLAIDKVY NPIGIVLMAR YMSKNYDFSK IPLAL LNPY DLTEFESVVK GWGETVNDRF LEVDNDAQRL IREKNILPED ILPDSLFSFR HVDVLLKRLF PRDPVSSFY UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: RNA (5'-R(P*AP*GP*UP*AP*GP*UP*AP*GP*AP*CP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*AP*GP*UP*AP*GP*UP*AP*GP*AP*CP*U)-3') / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Sin Nombre orthohantavirus Sin Nombre orthohantavirus |
| Molecular weight | Theoretical: 5.796515 KDa |
| Sequence | String: UAGUAGUAGA CUCCGAGA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)