[English] 日本語
 Yorodumi
Yorodumi- EMDB-16640: PfRH5-PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PfRH5-PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to antibody Cy.003 - map with additional PfRIPR tail density | |||||||||
 Map data Map data | map with additional PfRIPR tail density | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | malaria / erythrocyte invasion / Plasmodium falciparum / CELL ADHESION | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.95 Å | |||||||||
 Authors Authors | Farrell B / Higgins MK | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: The PfRCR complex bridges malaria parasite and erythrocyte during invasion. Authors: Brendan Farrell / Nawsad Alam / Melissa N Hart / Abhishek Jamwal / Robert J Ragotte / Hannah Walters-Morgan / Simon J Draper / Ellen Knuepfer / Matthew K Higgins /  Abstract: The symptoms of malaria occur during the blood stage of infection, when parasites invade and replicate within human erythrocytes. The PfPCRCR complex, containing PfRH5 (refs. ), PfCyRPA, PfRIPR, ...The symptoms of malaria occur during the blood stage of infection, when parasites invade and replicate within human erythrocytes. The PfPCRCR complex, containing PfRH5 (refs. ), PfCyRPA, PfRIPR, PfCSS and PfPTRAMP, is essential for erythrocyte invasion by the deadliest human malaria parasite, Plasmodium falciparum. Invasion can be prevented by antibodies or nanobodies against each of these conserved proteins, making them the leading blood-stage malaria vaccine candidates. However, little is known about how PfPCRCR functions during invasion. Here we present the structure of the PfRCR complex, containing PfRH5, PfCyRPA and PfRIPR, determined by cryogenic-electron microscopy. We test the hypothesis that PfRH5 opens to insert into the membrane, instead showing that a rigid, disulfide-locked PfRH5 can mediate efficient erythrocyte invasion. We show, through modelling and an erythrocyte-binding assay, that PfCyRPA-binding antibodies neutralize invasion through a steric mechanism. We determine the structure of PfRIPR, showing that it consists of an ordered, multidomain core flexibly linked to an elongated tail. We also show that the elongated tail of PfRIPR, which is the target of growth-neutralizing antibodies, binds to the PfCSS-PfPTRAMP complex on the parasite membrane. A modular PfRIPR is therefore linked to the merozoite membrane through an elongated tail, and its structured core presents PfCyRPA and PfRH5 to interact with erythrocyte receptors. This provides fresh insight into the molecular mechanism of erythrocyte invasion and opens the way to new approaches in rational vaccine design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16640.map.gz emd_16640.map.gz | 298.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16640-v30.xml emd-16640-v30.xml emd-16640.xml emd-16640.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16640.png emd_16640.png | 44.2 KB | ||
| Masks |  emd_16640_msk_1.map emd_16640_msk_1.map | 600.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16640.cif.gz emd-16640.cif.gz | 4 KB | ||
| Others |  emd_16640_half_map_1.map.gz emd_16640_half_map_1.map.gz emd_16640_half_map_2.map.gz emd_16640_half_map_2.map.gz | 556.9 MB 557 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16640 http://ftp.pdbj.org/pub/emdb/structures/EMD-16640 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16640 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16640 | HTTPS FTP |
-Validation report
| Summary document |  emd_16640_validation.pdf.gz emd_16640_validation.pdf.gz | 906.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16640_full_validation.pdf.gz emd_16640_full_validation.pdf.gz | 905.7 KB | Display | |
| Data in XML |  emd_16640_validation.xml.gz emd_16640_validation.xml.gz | 19.5 KB | Display | |
| Data in CIF |  emd_16640_validation.cif.gz emd_16640_validation.cif.gz | 23 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16640 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16640 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16640 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16640 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16640.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16640.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map with additional PfRIPR tail density | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
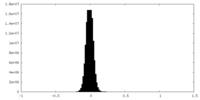
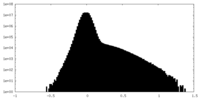
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16640_msk_1.map emd_16640_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
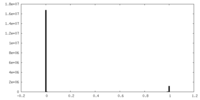
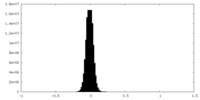
| Density Histograms |
-Half map: map with additional PfRIPR tail density - half map A
| File | emd_16640_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map with additional PfRIPR tail density - half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
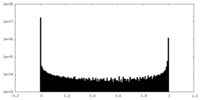
| Density Histograms |
-Half map: map with additional PfRIPR tail density - half map B
| File | emd_16640_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map with additional PfRIPR tail density - half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
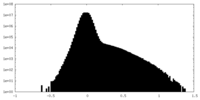
| Density Histograms |
- Sample components
Sample components
-Entire : PfRH5-PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to ...
| Entire | Name: PfRH5-PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to antibody Cy.003 |
|---|---|
| Components |
|
-Supramolecule #1: PfRH5-PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to ...
| Supramolecule | Name: PfRH5-PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to antibody Cy.003 type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 48.97 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.95 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 62817 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)