[English] 日本語
 Yorodumi
Yorodumi- EMDB-16471: in vitro reconstituted yeast 40S ribosome complex with deubiquiti... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | in vitro reconstituted yeast 40S ribosome complex with deubiquitinating enzyme Otu2 bound to ubiquitinated eS7 - body 2 (40S head) | ||||||||||||||||||
 Map data Map data | Cryo-EM map of in vitro reconstituted yeast 40S ribosome complex with deubiquitinating enzyme Otu2 bound to ubiquitinated eS7 - body 2 (40S head) | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
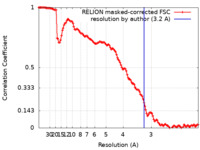
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||||||||
 Authors Authors | Ikeuchi K / Buschauer R / Cheng J / Berninghausen O / Becker T / Beckmann R | ||||||||||||||||||
| Funding support |  Germany, Germany,  Japan, 5 items Japan, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2. Authors: Ken Ikeuchi / Nives Ivic / Robert Buschauer / Jingdong Cheng / Thomas Fröhlich / Yoshitaka Matsuo / Otto Berninghausen / Toshifumi Inada / Thomas Becker / Roland Beckmann /     Abstract: In actively translating 80S ribosomes the ribosomal protein eS7 of the 40S subunit is monoubiquitinated by the E3 ligase Not4 and deubiquitinated by Otu2 upon ribosomal subunit recycling. Despite its ...In actively translating 80S ribosomes the ribosomal protein eS7 of the 40S subunit is monoubiquitinated by the E3 ligase Not4 and deubiquitinated by Otu2 upon ribosomal subunit recycling. Despite its importance for translation efficiency the exact role and structural basis for this translational reset is poorly understood. Here, structural analysis by cryo-electron microscopy of native and reconstituted Otu2-bound ribosomal complexes reveals that Otu2 engages 40S subunits mainly between ribosome recycling and initiation stages. Otu2 binds to several sites on the intersubunit surface of the 40S that are not occupied by any other 40S-binding factors. This binding mode explains the discrimination against 80S ribosomes via the largely helical N-terminal domain of Otu2 as well as the specificity for mono-ubiquitinated eS7 on 40S. Collectively, this study reveals mechanistic insights into the Otu2-driven deubiquitination steps for translational reset during ribosome recycling/(re)initiation. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16471.map.gz emd_16471.map.gz | 105.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16471-v30.xml emd-16471-v30.xml emd-16471.xml emd-16471.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16471_fsc.xml emd_16471_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_16471.png emd_16471.png | 70.1 KB | ||
| Others |  emd_16471_half_map_1.map.gz emd_16471_half_map_1.map.gz emd_16471_half_map_2.map.gz emd_16471_half_map_2.map.gz | 118.4 MB 118.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16471 http://ftp.pdbj.org/pub/emdb/structures/EMD-16471 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16471 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16471 | HTTPS FTP |
-Validation report
| Summary document |  emd_16471_validation.pdf.gz emd_16471_validation.pdf.gz | 867.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16471_full_validation.pdf.gz emd_16471_full_validation.pdf.gz | 866.8 KB | Display | |
| Data in XML |  emd_16471_validation.xml.gz emd_16471_validation.xml.gz | 20.3 KB | Display | |
| Data in CIF |  emd_16471_validation.cif.gz emd_16471_validation.cif.gz | 26.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16471 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16471 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16471 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16471 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16471.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16471.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of in vitro reconstituted yeast 40S ribosome complex with deubiquitinating enzyme Otu2 bound to ubiquitinated eS7 - body 2 (40S head) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||
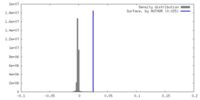
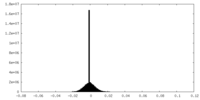
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map B
| File | emd_16471_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
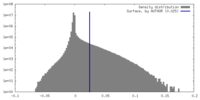
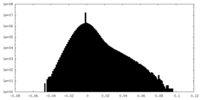
| Density Histograms |
-Half map: Half map A
| File | emd_16471_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : in vitro reconstituted yeast 40S ribosome complex with deubiquiti...
| Entire | Name: in vitro reconstituted yeast 40S ribosome complex with deubiquitinating enzyme Otu2 bound to ubiquitinated eS7 - body 2 (40S head) |
|---|---|
| Components |
|
-Supramolecule #1: in vitro reconstituted yeast 40S ribosome complex with deubiquiti...
| Supramolecule | Name: in vitro reconstituted yeast 40S ribosome complex with deubiquitinating enzyme Otu2 bound to ubiquitinated eS7 - body 2 (40S head) type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#18 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 46.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)