+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | GRM3C BMC shell from R. palustris, T=7 | |||||||||
 Map data Map data | Post-processed (sharpened, filtered) cryo-EM map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacterial microcompartment / BMC shell / GRM3 / STRUCTURAL PROTEIN | |||||||||
| Biological species |  Rhodopseudomonas palustris BisB18 (phototrophic) Rhodopseudomonas palustris BisB18 (phototrophic) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Greber BJ / Ferlez BH / Kirst H / Sutter M / Nogales E / Kerfeld CA | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Adv Mater / Year: 2023 Journal: Adv Mater / Year: 2023Title: Heterologous Assembly of Pleomorphic Bacterial Microcompartment Shell Architectures Spanning the Nano- to Microscale. Authors: Bryan H Ferlez / Henning Kirst / Basil J Greber / Eva Nogales / Markus Sutter / Cheryl A Kerfeld /  Abstract: Many bacteria use protein-based organelles known as bacterial microcompartments (BMCs) to organize and sequester sequential enzymatic reactions. Regardless of their specialized metabolic function, ...Many bacteria use protein-based organelles known as bacterial microcompartments (BMCs) to organize and sequester sequential enzymatic reactions. Regardless of their specialized metabolic function, all BMCs are delimited by a shell made of multiple structurally redundant, yet functionally diverse, hexameric (BMC-H), pseudohexameric/trimeric (BMC-T), or pentameric (BMC-P) shell protein paralogs. When expressed without their native cargo, shell proteins have been shown to self-assemble into 2D sheets, open-ended nanotubes, and closed shells of ≈40 nm diameter that are being developed as scaffolds and nanocontainers for applications in biotechnology. Here, by leveraging a strategy for affinity-based purification, it is demonstrated that a wide range of empty synthetic shells, many differing in end-cap structures, can be derived from a glycyl radical enzyme-associated microcompartment. The range of pleomorphic shells observed, which span ≈2 orders of magnitude in size from ≈25 nm to ≈1.8 µm, reveal the remarkable plasticity of BMC-based biomaterials. In addition, new capped nanotube and nanocone morphologies are observed that are consistent with a multicomponent geometric model in which architectural principles are shared among asymmetric carbon, viral protein, and BMC-based structures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16402.map.gz emd_16402.map.gz | 264.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16402-v30.xml emd-16402-v30.xml emd-16402.xml emd-16402.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16402.png emd_16402.png | 287.3 KB | ||
| Filedesc metadata |  emd-16402.cif.gz emd-16402.cif.gz | 4.7 KB | ||
| Others |  emd_16402_half_map_1.map.gz emd_16402_half_map_1.map.gz emd_16402_half_map_2.map.gz emd_16402_half_map_2.map.gz | 225.8 MB 226.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16402 http://ftp.pdbj.org/pub/emdb/structures/EMD-16402 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16402 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16402 | HTTPS FTP |
-Validation report
| Summary document |  emd_16402_validation.pdf.gz emd_16402_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16402_full_validation.pdf.gz emd_16402_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_16402_validation.xml.gz emd_16402_validation.xml.gz | 16.5 KB | Display | |
| Data in CIF |  emd_16402_validation.cif.gz emd_16402_validation.cif.gz | 19.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16402 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16402 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16402 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16402 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16402.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16402.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed (sharpened, filtered) cryo-EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.403 Å | ||||||||||||||||||||||||||||||||||||
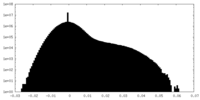
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Unfiltered half-map
| File | emd_16402_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
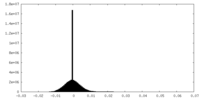
| Density Histograms |
-Half map: Unfiltered half-map
| File | emd_16402_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
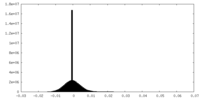
| Density Histograms |
- Sample components
Sample components
-Entire : Bacterial microcompartment shell from Rhodopseudomonas palustris ...
| Entire | Name: Bacterial microcompartment shell from Rhodopseudomonas palustris BisB18 |
|---|---|
| Components |
|
-Supramolecule #1: Bacterial microcompartment shell from Rhodopseudomonas palustris ...
| Supramolecule | Name: Bacterial microcompartment shell from Rhodopseudomonas palustris BisB18 type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Rhodopseudomonas palustris BisB18 (phototrophic) Rhodopseudomonas palustris BisB18 (phototrophic) |
| Molecular weight | Theoretical: 4 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - #0 - Film type ID: 3 / Support film - #0 - Material: GOLD / Support film - #0 - topology: HOLEY / Support film - #1 - Film type ID: 4 / Support film - #1 - Material: CARBON / Support film - #1 - topology: CONTINUOUS / Pretreatment - Type: PLASMA CLEANING | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 (6k x 4k) / #0 - Number real images: 378 / #0 - Average electron dose: 25.0 e/Å2 #0 - Details: Images collected on carbon-coated holey carbon grid. #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 (6k x 4k) / #1 - Number real images: 724 / #1 - Average electron dose: 25.0 e/Å2 #1 - Details: Images collected on carbon-coated holey gold grid. |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 35638 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)