+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | African cichlid nackednavirus capsid at pH 5.5 | ||||||||||||
 Map data Map data | Refined sharpened map of recombinant African Cichlid Nackednavirus capsid at pH 5.5 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | T=3 / Capsid protein / VIRAL PROTEIN | ||||||||||||
| Function / homology | C protein Function and homology information Function and homology information | ||||||||||||
| Biological species |  African cichlid nackednavirus African cichlid nackednavirus | ||||||||||||
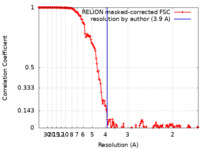
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | ||||||||||||
 Authors Authors | Pfister S / Rabl J / Boehringer D / Meier BH | ||||||||||||
| Funding support | European Union,  Switzerland, 3 items Switzerland, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural conservation of HBV-like capsid proteins over hundreds of millions of years despite the shift from non-enveloped to enveloped life-style. Authors: Sara Pfister / Julius Rabl / Thomas Wiegand / Simone Mattei / Alexander A Malär / Lauriane Lecoq / Stefan Seitz / Ralf Bartenschlager / Anja Böckmann / Michael Nassal / Daniel Boehringer / Beat H Meier /    Abstract: The discovery of nackednaviruses provided new insight into the evolutionary history of the hepatitis B virus (HBV): The common ancestor of HBV and nackednaviruses was non-enveloped and while HBV ...The discovery of nackednaviruses provided new insight into the evolutionary history of the hepatitis B virus (HBV): The common ancestor of HBV and nackednaviruses was non-enveloped and while HBV acquired an envelope during evolution, nackednaviruses remained non-enveloped. We report the capsid structure of the African cichlid nackednavirus (ACNDV), determined by cryo-EM at 3.7 Å resolution. This enables direct comparison with the known capsid structures of HBV and duck HBV, prototypic representatives of the mammalian and avian lineages of the enveloped Hepadnaviridae, respectively. The sequence identity with HBV is 24% and both the ACNDV capsid protein fold and the capsid architecture are very similar to those of the Hepadnaviridae and HBV in particular. Acquisition of the hepadnaviral envelope was thus not accompanied by a major change in capsid structure. Dynamic residues at the spike tip are tentatively assigned by solid-state NMR, while the C-terminal domain is invisible due to dynamics. Solid-state NMR characterization of the capsid structure reveals few conformational differences between the quasi-equivalent subunits of the ACNDV capsid and an overall higher capsid structural disorder compared to HBV. Despite these differences, the capsids of ACNDV and HBV are structurally highly similar despite the 400 million years since their separation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16371.map.gz emd_16371.map.gz | 206.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16371-v30.xml emd-16371-v30.xml emd-16371.xml emd-16371.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16371_fsc.xml emd_16371_fsc.xml | 15.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_16371.png emd_16371.png | 194.6 KB | ||
| Filedesc metadata |  emd-16371.cif.gz emd-16371.cif.gz | 6.5 KB | ||
| Others |  emd_16371_half_map_1.map.gz emd_16371_half_map_1.map.gz emd_16371_half_map_2.map.gz emd_16371_half_map_2.map.gz | 244.9 MB 244.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16371 http://ftp.pdbj.org/pub/emdb/structures/EMD-16371 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16371 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16371 | HTTPS FTP |
-Validation report
| Summary document |  emd_16371_validation.pdf.gz emd_16371_validation.pdf.gz | 874.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16371_full_validation.pdf.gz emd_16371_full_validation.pdf.gz | 873.8 KB | Display | |
| Data in XML |  emd_16371_validation.xml.gz emd_16371_validation.xml.gz | 23.2 KB | Display | |
| Data in CIF |  emd_16371_validation.cif.gz emd_16371_validation.cif.gz | 30.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16371 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16371 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16371 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16371 | HTTPS FTP |
-Related structure data
| Related structure data |  8c0oMC  8aacC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16371.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16371.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refined sharpened map of recombinant African Cichlid Nackednavirus capsid at pH 5.5 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
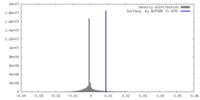
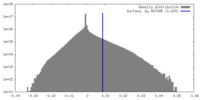
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map of recombinant African Cichlid Nackednavirus capsid...
| File | emd_16371_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of recombinant African Cichlid Nackednavirus capsid at pH 5.5 | ||||||||||||
| Projections & Slices |
| ||||||||||||
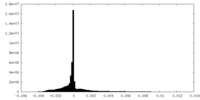
| Density Histograms |
-Half map: Half map of recombinant African Cichlid Nackednavirus capsid...
| File | emd_16371_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of recombinant African Cichlid Nackednavirus capsid at pH 5.5 | ||||||||||||
| Projections & Slices |
| ||||||||||||
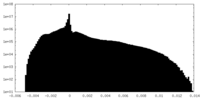
| Density Histograms |
- Sample components
Sample components
-Entire : African cichlid nackednavirus
| Entire | Name:  African cichlid nackednavirus African cichlid nackednavirus |
|---|---|
| Components |
|
-Supramolecule #1: African cichlid nackednavirus
| Supramolecule | Name: African cichlid nackednavirus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2497433 / Sci species name: African cichlid nackednavirus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Ophthalmotilapia ventralis (fish) Ophthalmotilapia ventralis (fish) |
| Molecular weight | Theoretical: 3.57 MDa |
| Virus shell | Shell ID: 1 / Name: capsid / Diameter: 230.0 Å / T number (triangulation number): 3 |
-Macromolecule #1: C protein
| Macromolecule | Name: C protein / type: protein_or_peptide / ID: 1 / Number of copies: 180 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  African cichlid nackednavirus African cichlid nackednavirus |
| Molecular weight | Theoretical: 19.851234 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGTFIELVKN MKGYKELLLP MEMVPLPAVV LKHVKLILTS QKEHQPWMTE MALKADQCLI HKATLDLAGK ATSNEAKPLI EAMQQIILA MTRELWGQIQ RHHYGIVQVE HYVKQITLWQ DTPQAFRGDQ PKPPSFRSDG PTRGQGSFRP FFRGRGRGRG R GRGSQSPA RKGPLPK UniProtKB: C protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 5.5 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 1 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | The protein concentration is approx. 0.1-0.5 mg/mL |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Frames/image: 1-40 / Number real images: 5362 / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 2.0 µm / Calibrated defocus min: 0.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 166000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | One protein chain was manually built in Coot. The chain was multiplied to give 180 chains, which were arranged as an icosahedral capsid. The capsid was refined with phenix and Isolde. |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-8c0o: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)