+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Enp1TAP_B, multibody refinement | |||||||||
 Map data Map data | Enp1TAP_B, multibody refinement | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Milkereit P / Poell G | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS One / Year: 2023 Journal: PLoS One / Year: 2023Title: Impact of the yeast S0/uS2-cluster ribosomal protein rpS21/eS21 on rRNA folding and the architecture of small ribosomal subunit precursors. Authors: Gisela Pöll / Joachim Griesenbeck / Herbert Tschochner / Philipp Milkereit /  Abstract: RpS0/uS2, rpS2/uS5, and rpS21/eS21 form a cluster of ribosomal proteins (S0-cluster) at the head-body junction near the central pseudoknot of eukaryotic small ribosomal subunits (SSU). Previous work ...RpS0/uS2, rpS2/uS5, and rpS21/eS21 form a cluster of ribosomal proteins (S0-cluster) at the head-body junction near the central pseudoknot of eukaryotic small ribosomal subunits (SSU). Previous work in yeast indicated that S0-cluster assembly is required for the stabilisation and maturation of SSU precursors at specific post-nucleolar stages. Here, we analysed the role of S0-cluster formation for rRNA folding. Structures of SSU precursors isolated from yeast S0-cluster expression mutants or control strains were analysed by cryogenic electron microscopy. The obtained resolution was sufficient to detect individual 2'-O-methyl RNA modifications using an unbiased scoring approach. The data show how S0-cluster formation enables the initial recruitment of the pre-rRNA processing factor Nob1 in yeast. Furthermore, they reveal hierarchical effects on the pre-rRNA folding pathway, including the final maturation of the central pseudoknot. Based on these structural insights we discuss how formation of the S0-cluster determines at this early cytoplasmic assembly checkpoint if SSU precursors further mature or are degraded. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16350.map.gz emd_16350.map.gz | 14.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16350-v30.xml emd-16350-v30.xml emd-16350.xml emd-16350.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16350.png emd_16350.png | 162 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16350 http://ftp.pdbj.org/pub/emdb/structures/EMD-16350 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16350 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16350 | HTTPS FTP |
-Validation report
| Summary document |  emd_16350_validation.pdf.gz emd_16350_validation.pdf.gz | 406.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16350_full_validation.pdf.gz emd_16350_full_validation.pdf.gz | 406.1 KB | Display | |
| Data in XML |  emd_16350_validation.xml.gz emd_16350_validation.xml.gz | 7.1 KB | Display | |
| Data in CIF |  emd_16350_validation.cif.gz emd_16350_validation.cif.gz | 8.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16350 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16350 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16350 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16350 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16350.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16350.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Enp1TAP_B, multibody refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.968 Å | ||||||||||||||||||||||||||||||||||||
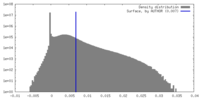
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Enp1-TAP associated immature ribosomal particles from S. cerevisiae
| Entire | Name: Enp1-TAP associated immature ribosomal particles from S. cerevisiae |
|---|---|
| Components |
|
-Supramolecule #1: Enp1-TAP associated immature ribosomal particles from S. cerevisiae
| Supramolecule | Name: Enp1-TAP associated immature ribosomal particles from S. cerevisiae type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#31 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 200 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 8575 / Average exposure time: 4.4 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
- Image processing
Image processing
| Final reconstruction | Number classes used: 1 / Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 4.0) / Number images used: 24592 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 4.0) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 4.0) |
| Final 3D classification | Software - Name: RELION (ver. 4.0) |
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)