+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Saccharolobus solfataricus type III-D CRISPR effector body 2 | |||||||||
 Map data Map data | Body 2 full map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Sulfolobus / CRISPR / Type III-D / ANTIVIRAL PROTEIN | |||||||||
| Biological species |   Saccharolobus solfataricus (archaea) Saccharolobus solfataricus (archaea) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Spagnolo L / White MF | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Curr Res Struct Biol / Year: 2023 Journal: Curr Res Struct Biol / Year: 2023Title: Structure of the type III-D CRISPR effector. Authors: Giuseppe Cannone / Dmytro Kompaniiets / Shirley Graham / Malcolm F White / Laura Spagnolo /  Abstract: CRISPR-Cas is a prokaryotic adaptive immune system, classified into six different types, each characterised by a signature protein. Type III systems, classified based on the presence of a Cas10 ...CRISPR-Cas is a prokaryotic adaptive immune system, classified into six different types, each characterised by a signature protein. Type III systems, classified based on the presence of a Cas10 subunit, are rather diverse multi-subunit assemblies with a range of enzymatic activities and downstream ancillary effectors. The broad array of current biotechnological CRISPR applications is mainly based on proteins classified as Type II, however recent developments established the feasibility and efficacy of multi-protein Type III CRISPR-Cas effector complexes as RNA-targeting tools in eukaryotes. The crenarchaeon has two type III system subtypes (III-B and III-D). Here, we report the cryo-EM structure of the Csm Type III-D complex from (SsoCsm), which uses CRISPR RNA to bind target RNA molecules, activating the Cas10 subunit for antiviral defence. The structure reveals the complex organisation, subunit/subunit connectivity and protein/guide RNA interactions of the SsoCsm complex, one of the largest CRISPR effectors known. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16175.map.gz emd_16175.map.gz | 78.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16175-v30.xml emd-16175-v30.xml emd-16175.xml emd-16175.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16175.png emd_16175.png | 63.6 KB | ||
| Filedesc metadata |  emd-16175.cif.gz emd-16175.cif.gz | 3.7 KB | ||
| Others |  emd_16175_half_map_1.map.gz emd_16175_half_map_1.map.gz emd_16175_half_map_2.map.gz emd_16175_half_map_2.map.gz | 78.4 MB 78.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16175 http://ftp.pdbj.org/pub/emdb/structures/EMD-16175 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16175 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16175 | HTTPS FTP |
-Validation report
| Summary document |  emd_16175_validation.pdf.gz emd_16175_validation.pdf.gz | 864.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16175_full_validation.pdf.gz emd_16175_full_validation.pdf.gz | 864.4 KB | Display | |
| Data in XML |  emd_16175_validation.xml.gz emd_16175_validation.xml.gz | 13.3 KB | Display | |
| Data in CIF |  emd_16175_validation.cif.gz emd_16175_validation.cif.gz | 15.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16175 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16175 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16175 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16175 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16175.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16175.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Body 2 full map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.046 Å | ||||||||||||||||||||||||||||||||||||
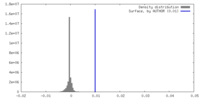
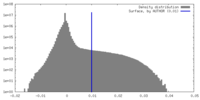
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Body 2 half map 1
| File | emd_16175_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Body 2 half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
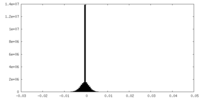
| Density Histograms |
-Half map: Body 2 half map 2
| File | emd_16175_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Body 2 half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
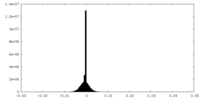
| Density Histograms |
- Sample components
Sample components
-Entire : Body 1 - Saccharolobus solfataricus type III-D CRISPR effector
| Entire | Name: Body 1 - Saccharolobus solfataricus type III-D CRISPR effector |
|---|---|
| Components |
|
-Supramolecule #1: Body 1 - Saccharolobus solfataricus type III-D CRISPR effector
| Supramolecule | Name: Body 1 - Saccharolobus solfataricus type III-D CRISPR effector type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Saccharolobus solfataricus (archaea) Saccharolobus solfataricus (archaea) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Average electron dose: 34.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.0 µm / Nominal defocus min: 0.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 192787 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)