[English] 日本語
 Yorodumi
Yorodumi- EMDB-1597: 5-fold averaged density map of Paramecium bursaria Chlorella viru... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1597 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
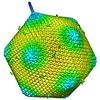
| Title | 5-fold averaged density map of Paramecium bursaria Chlorella virus-1 (PBCV-1). | |||||||||
 Map data Map data | 5-fold average density map of Paramecium bursaria Chlorella virus-1 (PBCV-1). | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PBCV-1 / eukaryotic virus / tail / pocket / 5-fold averaged | |||||||||
| Biological species |   Paramecium bursaria Chlorella virus 1 Paramecium bursaria Chlorella virus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 22.0 Å | |||||||||
 Authors Authors | Cherrier MV / Kostyuchenko VA / Xiao C / Bowman VD / Battisti AJ / Yan X / Chipman PR / Baker TS / Van Etten JL / Rossmann MG | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2009 Journal: Proc Natl Acad Sci U S A / Year: 2009Title: An icosahedral algal virus has a complex unique vertex decorated by a spike. Authors: Mickaël V Cherrier / Victor A Kostyuchenko / Chuan Xiao / Valorie D Bowman / Anthony J Battisti / Xiaodong Yan / Paul R Chipman / Timothy S Baker / James L Van Etten / Michael G Rossmann /  Abstract: Paramecium bursaria Chlorella virus-1 is an icosahedrally shaped, 1,900-A-diameter virus that infects unicellular eukaryotic green algae. A 5-fold symmetric, 3D reconstruction using cryoelectron ...Paramecium bursaria Chlorella virus-1 is an icosahedrally shaped, 1,900-A-diameter virus that infects unicellular eukaryotic green algae. A 5-fold symmetric, 3D reconstruction using cryoelectron microscopy images has now shown that the quasiicosahedral virus has a unique vertex, with a pocket on the inside and a spike structure on the outside of the capsid. The pocket might contain enzymes for use in the initial stages of infection. The unique vertex consists of virally coded proteins, some of which have been identified. Comparison of shape, size, and location of the spike with similar features in bacteriophages T4 and P22 suggests that the spike might be a cell-puncturing device. Similar asymmetric features may have been missed in previous analyses of many other viruses that had been assumed to be perfectly icosahedral. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1597.map.gz emd_1597.map.gz | 16.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1597-v30.xml emd-1597-v30.xml emd-1597.xml emd-1597.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-1597-image.png EMD-1597-image.png | 374.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1597 http://ftp.pdbj.org/pub/emdb/structures/EMD-1597 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1597 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1597 | HTTPS FTP |
-Validation report
| Summary document |  emd_1597_validation.pdf.gz emd_1597_validation.pdf.gz | 251.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1597_full_validation.pdf.gz emd_1597_full_validation.pdf.gz | 250.9 KB | Display | |
| Data in XML |  emd_1597_validation.xml.gz emd_1597_validation.xml.gz | 6.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1597 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1597 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1597 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1597 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1597.map.gz / Format: CCP4 / Size: 173.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1597.map.gz / Format: CCP4 / Size: 173.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 5-fold average density map of Paramecium bursaria Chlorella virus-1 (PBCV-1). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
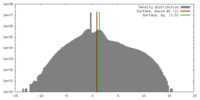
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Paramecium bursaria Chlorella virus-1 (PBCV-1)
| Entire | Name: Paramecium bursaria Chlorella virus-1 (PBCV-1) |
|---|---|
| Components |
|
-Supramolecule #1000: Paramecium bursaria Chlorella virus-1 (PBCV-1)
| Supramolecule | Name: Paramecium bursaria Chlorella virus-1 (PBCV-1) / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Paramecium bursaria Chlorella virus 1
| Supramolecule | Name: Paramecium bursaria Chlorella virus 1 / type: virus / ID: 1 / Name.synonym: PBCV-1 / NCBI-ID: 10506 / Sci species name: Paramecium bursaria Chlorella virus 1 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No / Syn species name: PBCV-1 |
|---|---|
| Host (natural) | Organism: Chlorella NC64A / synonym: ALGAE |
| Virus shell | Shell ID: 1 / Name: VP54 / Diameter: 1900 Å |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: Guillotine-style plunge freezing device Method: A small vial of ethane is placed inside a larger liquid nitrogen reservoir. The grid holding a few microliters of the sample is held in place at the bottom of a plunger by the means of fine ...Method: A small vial of ethane is placed inside a larger liquid nitrogen reservoir. The grid holding a few microliters of the sample is held in place at the bottom of a plunger by the means of fine tweezers. Once the ethane in the vial is completely frozen, it needs to be slightly melted. When the liquid ethane is ready, a piece of filter paper is then pressed against the sample to blot of excess buffer, sufficient to leave a thin layer on the grid. After a predetermined time, the filter paper is removed, and the plunger is allowed to drop into the liquid ethane. Once the grid enters the liquid ethane, the sample is rapidly frozen, and the grid is transferred under liquid nitrogen to a storage box immersed liquid nitrogen for later use in the microscope. |
|---|
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI/PHILIPS CM200FEG |
| Temperature | Average: 98 K |
| Alignment procedure | Legacy - Electron beam tilt params: 0 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7.0 µm / Number real images: 228 / Average electron dose: 22 e/Å2 Details: All particles were scaled to 4.0A per pixel, then bin twice to 8.0A per pixel. |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.473 µm / Nominal defocus min: 1.302 µm / Nominal magnification: 38000 |
| Sample stage | Specimen holder: EUCENTRIC / Specimen holder model: GATAN LIQUID NITROGEN |
- Electron microscopy #2
Electron microscopy #2
| Microscopy ID | 2 |
|---|---|
| Microscope | FEI/PHILIPS CM300FEG/T |
| Temperature | Average: 98 K |
| Alignment procedure | Legacy - Astigmatism: live FFT / Legacy - Electron beam tilt params: 0 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7.0 µm / Number real images: 228 / Average electron dose: 24 e/Å2 Details: All particles were scaled to 4.0A per pixel, then bin twice to 8.0A per pixel. |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 4.728 µm / Nominal defocus min: 0.767 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder: EUCENTRIC / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C5 (5 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 22.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Spider, XMIPP / Details: The 3D map was only 5 fold averaged. / Number images used: 5149 |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)