+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of cytochrome bd oxidase from C. glutamicum | |||||||||
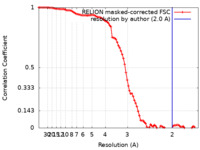
 Map data Map data | RELION masked postprocessing map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationcytochrome complex / aerobic electron transport chain / electron transfer activity / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Corynebacterium glutamicum (bacteria) Corynebacterium glutamicum (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.0 Å | |||||||||
 Authors Authors | Grund TN / Kusumoto T / Michel H / Sakamoto J / Safarian S | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: To be published Journal: To be publishedTitle: Cryo-EM structure of cytochrome bd oxidase from C. glutamicum Authors: Grund TN / Michel H / Safarian S | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15851.map.gz emd_15851.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15851-v30.xml emd-15851-v30.xml emd-15851.xml emd-15851.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15851_fsc.xml emd_15851_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_15851.png emd_15851.png | 59.9 KB | ||
| Masks |  emd_15851_msk_1.map emd_15851_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_15851_additional_1.map.gz emd_15851_additional_1.map.gz emd_15851_half_map_1.map.gz emd_15851_half_map_1.map.gz emd_15851_half_map_2.map.gz emd_15851_half_map_2.map.gz | 49.3 MB 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15851 http://ftp.pdbj.org/pub/emdb/structures/EMD-15851 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15851 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15851 | HTTPS FTP |
-Validation report
| Summary document |  emd_15851_validation.pdf.gz emd_15851_validation.pdf.gz | 705.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15851_full_validation.pdf.gz emd_15851_full_validation.pdf.gz | 705.1 KB | Display | |
| Data in XML |  emd_15851_validation.xml.gz emd_15851_validation.xml.gz | 14.3 KB | Display | |
| Data in CIF |  emd_15851_validation.cif.gz emd_15851_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15851 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15851 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15851 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15851 | HTTPS FTP |
-Related structure data
| Related structure data |  8b4oMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15851.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15851.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION masked postprocessing map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.837 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15851_msk_1.map emd_15851_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
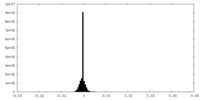
| Density Histograms |
-Additional map: RELION masked 3D refinement map
| File | emd_15851_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION masked 3D refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
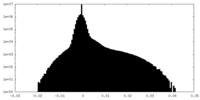
| Density Histograms |
-Half map: RELION 3D refinement half map 1
| File | emd_15851_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION 3D refinement half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
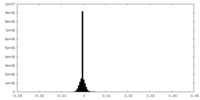
| Density Histograms |
-Half map: RELION 3D refinement half map 2
| File | emd_15851_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION 3D refinement half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
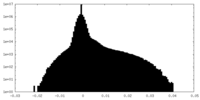
| Density Histograms |
- Sample components
Sample components
-Entire : CydAB heterodimer
| Entire | Name: CydAB heterodimer |
|---|---|
| Components |
|
-Supramolecule #1: CydAB heterodimer
| Supramolecule | Name: CydAB heterodimer / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Corynebacterium glutamicum (bacteria) Corynebacterium glutamicum (bacteria) |
| Molecular weight | Theoretical: 93 KDa |
-Macromolecule #1: Cytochrome bd-type quinol oxidase subunit II
| Macromolecule | Name: Cytochrome bd-type quinol oxidase subunit II / type: protein_or_peptide / ID: 1 / Details: cydB / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Corynebacterium glutamicum (bacteria) Corynebacterium glutamicum (bacteria) |
| Molecular weight | Theoretical: 36.261582 KDa |
| Recombinant expression | Organism:  Corynebacterium glutamicum (bacteria) Corynebacterium glutamicum (bacteria) |
| Sequence | String: MDHNTFWFIL IAFLFSGYFL LEGFDFGVGI LAPIIGKDSA ARNTVIRTIG PVWDGNEVWL IVAGGALFAA FPEWYATMFS GMYLPLFLV LVSLIIRVVG LEWRKKVDDP RWQKWSDRAI FIGSWTPPLM WGFIFANILR GMPIKADHTI DAAAALPGMV N VFAILGAL ...String: MDHNTFWFIL IAFLFSGYFL LEGFDFGVGI LAPIIGKDSA ARNTVIRTIG PVWDGNEVWL IVAGGALFAA FPEWYATMFS GMYLPLFLV LVSLIIRVVG LEWRKKVDDP RWQKWSDRAI FIGSWTPPLM WGFIFANILR GMPIKADHTI DAAAALPGMV N VFAILGAL AFTALFALHG LAFIRLKTAG RVRTDAAKAA PGVALLAAVT GGPFVLWAAI AYGRSWSWIL AVLIIAAVLG GA FALIKDR DGLSFLSTSV AVIGVVALLF SSLFPNVMPT TLADGVSLDI WNASASHYAL TILTWTAAVI APLVVLYQGW TYW VFRKRL HAEPVSA |
-Macromolecule #2: Cytochrome BD ubiquinol oxidase subunit I
| Macromolecule | Name: Cytochrome BD ubiquinol oxidase subunit I / type: protein_or_peptide / ID: 2 / Details: CydA / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Corynebacterium glutamicum (bacteria) Corynebacterium glutamicum (bacteria) |
| Molecular weight | Theoretical: 56.678461 KDa |
| Recombinant expression | Organism:  Corynebacterium glutamicum (bacteria) Corynebacterium glutamicum (bacteria) |
| Sequence | String: MDVVDIARWQ FGITTVYHFI FVPLTIGLAP LVAIMQTFWQ VTGKEHWYRA TRFFGTVLLI NFAVGVATGI VQEFQFGMNW SEYSRFVGD VFGGPLALEG LIAFFLESVF LGLWIFGWGK IPGWLHTASI WIVAIATNIS AYFIIVANSF MQHPVGAEYN P ETGRAELT ...String: MDVVDIARWQ FGITTVYHFI FVPLTIGLAP LVAIMQTFWQ VTGKEHWYRA TRFFGTVLLI NFAVGVATGI VQEFQFGMNW SEYSRFVGD VFGGPLALEG LIAFFLESVF LGLWIFGWGK IPGWLHTASI WIVAIATNIS AYFIIVANSF MQHPVGAEYN P ETGRAELT DFWALLTNST ALAAFPHAVA GGFLTAGTFV LGISGWWIIR AHRQAKKAEA EIESKHSMHR PALWVGWWTT VV SSVALFI TGDTQAKLMF VQQPMKMASA ESLCETATDP NFSILTIGTH NNCDTVTHLI DVPFVLPFLA EGKFTGVTLQ GVN QLQAAA EQAYGPGNYS PNLFVTYWSF RAMIGLMLGS LAIAAIAWLL LRKKRTPTGK IARLFQIGSL IAIPFPFLAN SAGW IFTEM GRQPWVVHPN PESAGDARTE MIRMTVDMGV SDHAPWQVWL TLIGFTILYL ILFVVWVWLI RRAVLIGPPE EGAPS VEAK TGPATPIGSD MPMTPLQFTA AAPTTREKE |
-Macromolecule #3: CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE
| Macromolecule | Name: CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE / type: ligand / ID: 3 / Number of copies: 1 / Formula: HDD |
|---|---|
| Molecular weight | Theoretical: 632.487 Da |
| Chemical component information |  ChemComp-HDD: |
-Macromolecule #4: HEME B/C
| Macromolecule | Name: HEME B/C / type: ligand / ID: 4 / Number of copies: 2 / Formula: HEB |
|---|---|
| Molecular weight | Theoretical: 618.503 Da |
| Chemical component information |  ChemComp-HEB: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 107.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 1.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z
Z Y
Y X
X