[English] 日本語
 Yorodumi
Yorodumi- EMDB-15651: Structure of the giant inhibitor of apoptosis, BIRC6 (multibody map 3) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the giant inhibitor of apoptosis, BIRC6 (multibody map 3) | |||||||||
 Map data Map data | Multibody map 3 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | E2/E3 ubiquitin ligase / APOPTOSIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationspongiotrophoblast layer development / labyrinthine layer development / ALK mutants bind TKIs / Flemming body / microtubule organizing center / cysteine-type endopeptidase inhibitor activity / ubiquitin conjugating enzyme activity / regulation of cytokinesis / negative regulation of extrinsic apoptotic signaling pathway / trans-Golgi network ...spongiotrophoblast layer development / labyrinthine layer development / ALK mutants bind TKIs / Flemming body / microtubule organizing center / cysteine-type endopeptidase inhibitor activity / ubiquitin conjugating enzyme activity / regulation of cytokinesis / negative regulation of extrinsic apoptotic signaling pathway / trans-Golgi network / RING-type E3 ubiquitin transferase / spindle pole / ubiquitin-protein transferase activity / Signaling by ALK fusions and activated point mutants / regulation of cell population proliferation / midbody / cell population proliferation / endosome / protein ubiquitination / protein phosphorylation / cell division / centrosome / positive regulation of cell population proliferation / negative regulation of apoptotic process / apoptotic process / membrane / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Dietz L / Elliott PR | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Structural basis for SMAC-mediated antagonism of caspase inhibition by the giant ubiquitin ligase BIRC6. Authors: Larissa Dietz / Cara J Ellison / Carlos Riechmann / C Keith Cassidy / F Daniel Felfoldi / Adán Pinto-Fernández / Benedikt M Kessler / Paul R Elliott /  Abstract: Certain inhibitor of apoptosis (IAP) family members are sentinel proteins that prevent untimely cell death by inhibiting caspases. Antagonists, including second mitochondria-derived activator of ...Certain inhibitor of apoptosis (IAP) family members are sentinel proteins that prevent untimely cell death by inhibiting caspases. Antagonists, including second mitochondria-derived activator of caspases (SMAC), regulate IAPs and drive cell death. Baculoviral IAP repeat-containing protein 6 (BIRC6), a giant IAP with dual E2 and E3 ubiquitin ligase activity, regulates programmed cell death through unknown mechanisms. We show that BIRC6 directly restricts executioner caspase-3 and -7 and ubiquitinates caspase-3, -7, and -9, working exclusively with noncanonical E1, UBA6. Notably, we show that SMAC suppresses both mechanisms. Cryo-electron microscopy structures of BIRC6 alone and in complex with SMAC reveal that BIRC6 is an antiparallel dimer juxtaposing the substrate-binding module against the catalytic domain. Furthermore, we discover that SMAC multisite binding to BIRC6 results in a subnanomolar affinity interaction, enabling SMAC to competitively displace caspases, thus antagonizing BIRC6 anticaspase function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15651.map.gz emd_15651.map.gz | 96.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15651-v30.xml emd-15651-v30.xml emd-15651.xml emd-15651.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15651.png emd_15651.png | 18.9 KB | ||
| Masks |  emd_15651_msk_1.map emd_15651_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15651.cif.gz emd-15651.cif.gz | 7.9 KB | ||
| Others |  emd_15651_half_map_1.map.gz emd_15651_half_map_1.map.gz emd_15651_half_map_2.map.gz emd_15651_half_map_2.map.gz | 56.7 MB 56.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15651 http://ftp.pdbj.org/pub/emdb/structures/EMD-15651 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15651 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15651 | HTTPS FTP |
-Validation report
| Summary document |  emd_15651_validation.pdf.gz emd_15651_validation.pdf.gz | 983.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15651_full_validation.pdf.gz emd_15651_full_validation.pdf.gz | 982.9 KB | Display | |
| Data in XML |  emd_15651_validation.xml.gz emd_15651_validation.xml.gz | 13 KB | Display | |
| Data in CIF |  emd_15651_validation.cif.gz emd_15651_validation.cif.gz | 15.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15651 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15651 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15651 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15651 | HTTPS FTP |
-Related structure data
| Related structure data |  8atmC  8atoC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15651.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15651.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Multibody map 3 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
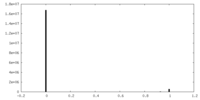
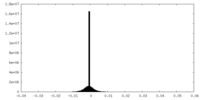
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15651_msk_1.map emd_15651_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
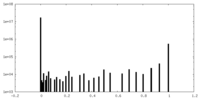
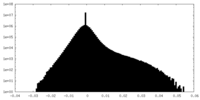
| Density Histograms |
-Half map: 2nd half map multibody map 3
| File | emd_15651_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 2nd half map multibody map 3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
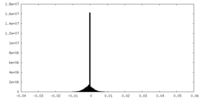
| Density Histograms |
-Half map: 1st half map multibody map 3
| File | emd_15651_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 1st half map multibody map 3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
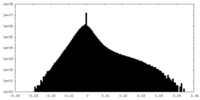
| Density Histograms |
- Sample components
Sample components
-Entire : Anti-parallel homodimer
| Entire | Name: Anti-parallel homodimer |
|---|---|
| Components |
|
-Supramolecule #1: Anti-parallel homodimer
| Supramolecule | Name: Anti-parallel homodimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1 MDa |
-Macromolecule #1: BIRC6
| Macromolecule | Name: BIRC6 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPMVTGGGAA PPGTVTEPLP SVIVLSAGRK MAAAAAAASG PGCSSAAGAG AAGVSEWLVL RDGCMHCDAD GLHSLSYHPA LNAILAVTSR GTIKVIDGTS GATLQASALS AKPGGQVKCQ YISAVDKVIF VDDYAVGCRK DLNGILLLDT ALQTPVSKQD DVVQLELPVT ...String: GPMVTGGGAA PPGTVTEPLP SVIVLSAGRK MAAAAAAASG PGCSSAAGAG AAGVSEWLVL RDGCMHCDAD GLHSLSYHPA LNAILAVTSR GTIKVIDGTS GATLQASALS AKPGGQVKCQ YISAVDKVIF VDDYAVGCRK DLNGILLLDT ALQTPVSKQD DVVQLELPVT EAQQLLSACL EKVDISSTEG YDLFITQLKD GLKNTSHETA ANHKVAKWAT VTFHLPHHVL KSIASAIVNE LKKINQNVAA LPVASSVMDR LSYLLPSARP ELGVGPGRSV DRSLMYSEAN RRETFTSWPH VGYRWAQPDP MAQAGFYHQP ASSGDDRAMC FTCSVCLVCW EPTDEPWSEH ERHSPNCPFV KGEHTQNVPL SVTLATSPAQ FPCTDGTDRI SCFGSGSCPH FLAAATKRGK ICIWDVSKLM KVHLKFEINA YDPAIVQQLI LSGDPSSGVD SRRPTLAWLE DSSSCSDIPK LEGDSDDLLE DSDSEEHSRS DSVTGHTSQK EAMEVSLDIT ALSILQQPEK LQWEIVANVL EDTVKDLEEL GANPCLTNSK SEKTKEKHQE QHNIPFPCLL AGGLLTYKSP ATSPISSNSH RSLDGLSRTQ GESISEQGST DNESCTNSEL NSPLVRRTLP VLLLYSIKES DEKAGKIFSQ MNNIMSKSLH DDGFTVPQII EMELDSQEQL LLQDPPVTYI QQFADAAANL TSPDSEKWNS VFPKPGTLVQ CLRLPKFAEE ENLCIDSITP CADGIHLLVG LRTCPVESLS AINQVEALNN LNKLNSALCN RRKGELESNL AVVNGANISV IQHESPADVQ TPLIIQPEQR NVSGGYLVLY KMNYATRIVT LEEEPIKIQH IKDPQDTITS LILLPPDILD NREDDCEEPI EDMQLTSKNG FEREKTSDIS TLGHLVITTQ GGYVKILDLS NFEILAKVEP PKKEGTEEQD TFVSVIYCSG TDRLCACTKG GELHFLQIGG TCDDIDEADI LVDGSLSKGI EPSSEGSKPL SNPSSPGISG VDLLVDQPFT LEILTSLVEL TRFETLTPRF SATVPPCWVE VQQEQQQRRH PQHLHQQHHG DAAQHTRTWK LQTDSNSWDE HVFELVLPKA CMVGHVDFKF VLNSNITNIP QIQVTLLKNK APGLGKVNAL NIEVEQNGKP SLVDLNEEMQ HMDVEESQCL RLCPFLEDHK EDILCGPVWL ASGLDLSGHA GMLTLTSPKL VKGMAGGKYR SFLIHVKAVN ERGTEEICNG GMRPVVRLPS LKHQSNKGYS LASLLAKVAA GKEKSSNVKN ENTSGTRKSE NLRGCDLLQE VSVTIRRFKK TSISKERVQR CAMLQFSEFH EKLLNTLCRK TDDGQITEHA QSLVLDTLCW LAGVHSNGPG SSKEGNENLL SKTRKFLSDI VRVCFFEAGR SIAHKCARFL ALCISNGKCD PCQPAFGPVL LKALLDNMSF LPAATTGGSV YWYFVLLNYV KDEDLAGCST ACASLLTAVS RQLQDRLTPM EALLQTRYGL YSSPFDPVLF DLEMSGSSCK NVYNSSIGVQ SDEIDLSDVL SGNGKVSSCT AAEGSFTSLT GLLEVEPLHF TCVSTSDGTR IERDDAMSSF GVTPAVGGLS SGTVGEASTA LSSAAQVALQ SLSHAMASAE QQLQVLQEKQ QQLLKLQQQK AKLEAKLHQT TAAAAAAASA VGPVHNSVPS NPVAAPGFFI HPSDVIPPTP KTTPLFMTPP LTPPNEAVSV VINAELAQLF PGSVIDPPAV NLAAHNKNSN KSRMNPLGSG LALAISHASH FLQPPPHQSI IIERMHSGAR RFVTLDFGRP ILLTDVLIPT CGDLASLSID IWTLGEEVDG RRLVVATDIS THSLILHDLI PPPVCRFMKI TVIGRYGSTN ARAKIPLGFY YGHTYILPWE SELKLMHDPL KGEGESANQP EIDQHLAMMV ALQEDIQCRY NLACHRLETL LQSIDLPPLN SANNAQYFLR KPDKAVEEDS RVFSAYQDCI QLQLQLNLAH NAVQRLKVAL GASRKMLSET SNPEDLIQTS STEQLRTIIR YLLDTLLSLL HASNGHSVPA VLQSTFHAQA CEELFKHLCI SGTPKIRLHT GLLLVQLCGG ERWWGQFLSN VLQELYNSEQ LLIFPQDRVF MLLSCIGQRS LSNSGVLESL LNLLDNLLSP LQPQLPMHRR TEGVLDIPMI SWVVMLVSRL LDYVATVEDE AAAAKKPLNG NQWSFINNNL HTQSLNRSSK GSSSLDRLYS RKIRKQLVHH KQQLNLLKAK QKALVEQMEK EKIQSNKGSS YKLLVEQAKL KQATSKHFKD LIRLRRTAEW SRSNLDTEVT TAKESPEIEP LPFTLAHERC ISVVQKLVLF LLSMDFTCHA DLLLFVCKVL ARIANATRPT IHLCEIVNEP QLERLLLLLV GTDFNRGDIS WGGAWAQYSL TCMLQDILAG ELLAPVAAEA MEEGTVGDDV GATAGDSDDS LQQSSVQLLE TIDEPLTHDI TGAPPLSSLE KDKEIDLELL QDLMEVDIDP LDIDLEKDPL AAKVFKPISS TWYDYWGADY GTYNYNPYIG GLGIPVAKPP ANTEKNGSQT VSVSVSQALD ARLEVGLEQQ AELMLKMMST LEADSILQAL TNTSPTLSQS PTGTDDSLLG GLQAANQTSQ LIIQLSSVPM LNVCFNKLFS MLQVHHVQLE SLLQLWLTLS LNSSSTGNKE NGADIFLYNA NRIPVISLNQ ASITSFLTVL AWYPNTLLRT WCLVLHSLTL MTNMQLNSGS SSAIGTQEST AHLLVSDPNL IHVLVKFLSG TSPHGTNQHS PQVGPTATQA MQEFLTRLQV HLSSTCPQIF SEFLLKLIHI LSTERGAFQT GQGPLDAQVK LLEFTLEQNF EVVSVSTISA VIESVTFLVH HYITCSDKVM SRSGSDSSVG ARACFGGLFA NLIRPGDAKA VCGEMTRDQL MFDLLKLVNI LVQLPLSGNR EYSARVSVTT NTTDSVSDEE KVSGGKDGNG SSTSVQGSPA YVADLVLANQ QIMSQILSAL GLCNSSAMAM IIGASGLHLT KHENFHGGLD AISVGDGLFT ILTTLSKKAS TVHMMLQPIL TYMACGYMGR QGSLATCQLS EPLLWFILRV LDTSDALKAF HDMGGVQLIC NNMVTSTRAI VNTARSMVST IMKFLDSGPN KAVDSTLKTR ILASEPDNAE GIHNFAPLGT ITSSSPTAQP AEVLLQATPP HRRARSAAWS YIFLPEEAWC DLTIHLPAAV LLKEIHIQPH LASLATCPSS VSVEVSADGV NMLPLSTPVV TSGLTYIKIQ LVKAEVASAV CLRLHRPRDA STLGLSQIKL LGLTAFGTTS SATVNNPFLP SEDQVSKTSI GWLRLLHHCL THISDLEGMM ASAAAPTANL LQTCAALLMS PYCGMHSPNI EVVLVKIGLQ STRIGLKLID ILLRNCAASG SDPTDLNSPL LFGRLNGLSS DSTIDILYQL GTTQDPGTKD RIQALLKWVS DSARVAAMKR SGRMNYMCPN SSTVEYGLLM PSPSHLHCVA AILWHSYELL VEYDLPALLD QELFELLFNW SMSLPCNMVL KKAVDSLLCS MCHVHPNYFS LLMGWMGITP PPVQCHHRLS MTDDSKKQDL SSSLTDDSKN AQAPLALTES HLATLASSSQ SPEAIKQLLD SGLPSLLVRS LASFCFSHIS SSESIAQSID ISQDKLRRHH VPQQCNKMPI TADLVAPILR FLTEVGNSHI MKDWLGGSEV NPLWTALLFL LCHSGSTSGS HNLGAQQTSA RSASLSSAAT TGLTTQQRTA IENATVAFFL QCISCHPNNQ KLMAQVLCEL FQTSPQRGNL PTSGNISGFI RRLFLQLMLE DEKVTMFLQS PCPLYKGRIN ATSHVIQHPM YGAGHKFRTL HLPVSTTLSD VLDRVSDTPS ITAKLISEQK DDKEKKNHEE KEKVKAENGF QDNYSVVVAS GLKSQSKRAV SATPPRPPSR RGRTIPDKIG STSGAEAANK IITVPVFHLF HKLLAGQPLP AEMTLAQLLT LLYDRKLPQG YRSIDLTVKL GSRVITDPSL SKTDSYKRLH PEKDHGDLLA SCPEDEALTP GDECMDGILD ESLLETCPIQ SPLQVFAGMG GLALIAERLP MLYPEVIQQV SAPVVTSTTQ EKPKDSDQFE WVTIEQSGEL VYEAPETVAA EPPPIKSAVQ TMSPIPAHSL AAFGLFLRLP GYAEVLLKER KHAQCLLRLV LGVTDDGEGS HILQSPSANV LPTLPFHVLR SLFSTTPLTT DDGVLLRRMA LEIGALHLIL VCLSALSHHS PRVPNSSVNQ TEPQVSSSHN PTSTEEQQLY WAKGTGFGTG STASGWDVEQ ALTKQRLEEE HVTCLLQVLA SYINPVSSAV NGEAQSSHET RGQNSNALPS VLLELLSQSC LIPAMSSYLR NDSVLDMARH VPLYRALLEL LRAIASCAAM VPLLLPLSTE NGEEEEEQSE CQTSVGTLLA KMKTCVDTYT NRLRSKRENV KTGVKPDASD QEPEGLTLLV PDIQKTAEIV YAATTSLRQA NQEKKLGEYS KKAAMKPKPL SVLKSLEEKY VAVMKKLQFD TFEMVSEDED GKLGFKVNYH YMSQVKNAND ANSAARARRL AQEAVTLSTS LPLSSSSSVF VRCDEERLDI MKVLITGPAD TPYANGCFEF DVYFPQDYPS SPPLVNLETT GGHSVRFNPN LYNDGKVCLS ILNTWHGRPE EKWNPQTSSF LQVLVSVQSL ILVAEPYFNE PGYERSRGTP SGTQSSREYD GNIRQATVKW AMLEQIRNPS PCFKEVIHKH FYLKRVEIMA QCEEWIADIQ QYSSDKRVGR TMSHHAAALK RHTAQLREEL LKLPCPEGLD PDTDDAPEVC RATTGAEETL MHDQVKPSSS KELPSDFQL UniProtKB: Baculoviral IAP repeat-containing protein 6 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 3.5 sec. / Average electron dose: 49.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 1.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)