+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human 80S ribosome structure from pFIB-lamellae | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
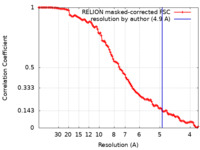
| Method | subtomogram averaging / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Berger C / Grange M | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Plasma FIB milling for the determination of structures in situ. Authors: Casper Berger / Maud Dumoux / Thomas Glen / Neville B-Y Yee / John M Mitchels / Zuzana Patáková / Michele C Darrow / James H Naismith / Michael Grange /   Abstract: Structural biology studies inside cells and tissues require methods to thin vitrified specimens to electron transparency. Until now, focused ion beams based on gallium have been used. However, ion ...Structural biology studies inside cells and tissues require methods to thin vitrified specimens to electron transparency. Until now, focused ion beams based on gallium have been used. However, ion implantation, changes to surface chemistry and an inability to access high currents limit gallium application. Here, we show that plasma-coupled ion sources can produce cryogenic lamellae of vitrified human cells in a robust and automated manner, with quality sufficient for pseudo-atomic structure determination. Lamellae were produced in a prototype microscope equipped for long cryogenic run times (> 1 week) and with multi-specimen support fully compatible with modern-day transmission electron microscopes. We demonstrate that plasma ion sources can be used for structural biology within cells, determining a structure in situ to 4.9 Å, and characterise the resolution dependence on particle distance from the lamella edge. We describe a workflow upon which different plasmas can be examined to further streamline lamella fabrication. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15636.map.gz emd_15636.map.gz | 12.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15636-v30.xml emd-15636-v30.xml emd-15636.xml emd-15636.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15636_fsc.xml emd_15636_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_15636.png emd_15636.png | 75.6 KB | ||
| Masks |  emd_15636_msk_1.map emd_15636_msk_1.map | 129.7 MB |  Mask map Mask map | |
| Others |  emd_15636_half_map_1.map.gz emd_15636_half_map_1.map.gz emd_15636_half_map_2.map.gz emd_15636_half_map_2.map.gz | 66.7 MB 66.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15636 http://ftp.pdbj.org/pub/emdb/structures/EMD-15636 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15636 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15636 | HTTPS FTP |
-Validation report
| Summary document |  emd_15636_validation.pdf.gz emd_15636_validation.pdf.gz | 819.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15636_full_validation.pdf.gz emd_15636_full_validation.pdf.gz | 818.9 KB | Display | |
| Data in XML |  emd_15636_validation.xml.gz emd_15636_validation.xml.gz | 18.7 KB | Display | |
| Data in CIF |  emd_15636_validation.cif.gz emd_15636_validation.cif.gz | 24.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15636 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15636 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15636 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15636 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15636.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15636.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.9 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15636_msk_1.map emd_15636_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15636_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15636_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 80S human ribosome
| Entire | Name: 80S human ribosome |
|---|---|
| Components |
|
-Supramolecule #1: 80S human ribosome
| Supramolecule | Name: 80S human ribosome / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 5.75 |
|---|---|
| Grid | Model: UltrAuFoil R2/2 / Material: GOLD / Mesh: 200 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV Details: 24 h post infection cells were plunge-frozen using the Vitrobot (Thermo Fisher) offsetting the blotting pad to favour back blotting. Just prior the PF, the cells media has been replaced with ...Details: 24 h post infection cells were plunge-frozen using the Vitrobot (Thermo Fisher) offsetting the blotting pad to favour back blotting. Just prior the PF, the cells media has been replaced with the complete media with 10% glycerol (v/v).. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 2 / Average electron dose: 5.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)