+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | E. coli Wadjet JetABC monomer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology | Wadjet protein JetB / Domain of unknown function (DUF4194) / Protein of unknown function DUF3375 / Protein of unknown function (DUF3375) / P-loop containing nucleoside triphosphate hydrolase / DUF4194 domain-containing protein / ATP synthase / DUF3375 family protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Roisne-Hamelin F / Beckert B / Li Y / Myasnikov A / Gruber S | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Molecular Cell / Year: 2022 Journal: Molecular Cell / Year: 2022Title: DNA-measuring Wadjet SMC ATPases restrict smaller circular plasmids by DNA cleavage. Authors: Liu HW / Roisne-Hamelin F / Beckert B / Li Y / Myasnikov A / Gruber S | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15609.map.gz emd_15609.map.gz | 10.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15609-v30.xml emd-15609-v30.xml emd-15609.xml emd-15609.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
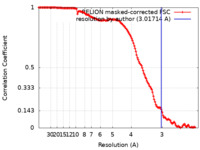
| FSC (resolution estimation) |  emd_15609_fsc.xml emd_15609_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_15609.png emd_15609.png | 83.3 KB | ||
| Masks |  emd_15609_msk_1.map emd_15609_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Others |  emd_15609_additional_1.map.gz emd_15609_additional_1.map.gz emd_15609_half_map_1.map.gz emd_15609_half_map_1.map.gz emd_15609_half_map_2.map.gz emd_15609_half_map_2.map.gz | 15.2 MB 194.3 MB 194.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15609 http://ftp.pdbj.org/pub/emdb/structures/EMD-15609 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15609 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15609 | HTTPS FTP |
-Validation report
| Summary document |  emd_15609_validation.pdf.gz emd_15609_validation.pdf.gz | 692.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15609_full_validation.pdf.gz emd_15609_full_validation.pdf.gz | 691.9 KB | Display | |
| Data in XML |  emd_15609_validation.xml.gz emd_15609_validation.xml.gz | 21.8 KB | Display | |
| Data in CIF |  emd_15609_validation.cif.gz emd_15609_validation.cif.gz | 28.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15609 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15609 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15609 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15609 | HTTPS FTP |
-Related structure data
| Related structure data |  8as8M M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15609.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15609.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1616 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15609_msk_1.map emd_15609_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_15609_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15609_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15609_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : JetABC(D)
| Entire | Name: JetABC(D) |
|---|---|
| Components |
|
-Supramolecule #1: JetABC(D)
| Supramolecule | Name: JetABC(D) / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#3 Details: E. coli JetABCD was purified by gel filtration. Note that JetD is not visible in the map. |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: JetC
| Macromolecule | Name: JetC / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 124.562938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNQVSGLAGK ESFILTRIEL FNWGGFHGLH QAAIHQDGTA VIGPTGSGKT TLVDALMTLL CANPRYNLAS TGGHESDRDL ISYVRGVSG PGDGGEGQSH IARPGKTVTG IAATLEREGK QVRLGALLWF DSTSSSVTDM KRLWLFSDNP GQTLEHWLNV Y HEGGTRLL ...String: MNQVSGLAGK ESFILTRIEL FNWGGFHGLH QAAIHQDGTA VIGPTGSGKT TLVDALMTLL CANPRYNLAS TGGHESDRDL ISYVRGVSG PGDGGEGQSH IARPGKTVTG IAATLEREGK QVRLGALLWF DSTSSSVTDM KRLWLFSDNP GQTLEHWLNV Y HEGGTRLL RQMEKEAIGL WTYPNKKQYL ARLRDFFEVG ENAFTLLNRA AGLKQLNSID EIFRELVLDD HSAFDRAAEV AN SFDGLTE IHQELETARK QQQSLQPVAL SWEKYQKQER QLADWLTLES LLPLWFAQQA SHLWREKINL LNARLAEAQT SEE QLQSQL DLQKKVVSDC MQRYLQVGGA NIDELNERIK DWQKTLGSRE ALARQYQQLT RNLGLPSDLS QPQLEANQHE AEAR CEQIA VDIKLKQEEA YQKGALSHHI TEELRERENE RAEIARRPDS NLPAHYQAFR SELAKALNVD ESELPFVAEL IQVKP EEAQ WRGAIERAVG SNRLRILVAP ESAQEALRWV NQRNNRLHVR LLEVKLPHSP ARFFDDGFTR KLLWKDHPWR EAVKAL LAE SDRHCVDSPE QLHDTPHAMT VQGLMSGKQR FYDKHDQKRL DEDWLTGFDN RDRLNFLAKE IATLQEQVKT ANAAFEF AK GEVGLLQNQA ASFQKIEQID FDSIDVPGAK SQLDALRERL ENLTRPDSDA SVAKAKLDEA QTIESELDKQ LRAANKVT N VLDTELTLAR AAERKAQQTA QQGMKEEERE LCASHFPVVT LEQLPDIRDL ERQHERGIQH EIERVKAELH RLNIELTKR MSEAKRVDTG ALVEAGADLD DIPVYLQRLQ ELTEEALPEK LNRFLDYLNR SSDDGVTQLL SHIEHEVLVI EERLNELNET MFRVDFQPD RYLRLDTKKV VHESLRTLEK AQRQLNAARF VDDNGESHYK ALQVLVAQLR DACERNRTLG AKALLDPRFR L EFAVSVMD RQSGNVIESR TGSQGGSGGE KEIIASYVLT ASLSYALCPA GSRYPLFGTI ILDEAFSRSS HAVAGRIIAA LR EFGLHAV FITPNKEMRL LRDHTRSAIV VHRRGQNSNM ASLSWEELER HYQRRGNAG |
-Macromolecule #2: JetB
| Macromolecule | Name: JetB / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.020416 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGFFDKLIN RSVTANAGCE PEPSDEEVTD ESVEDSLASS ETRTLQKIRE ATQELLKYGL LEEASKPNLY RIVLSHPEEV TRILEPLDL DIGIDEIRGL LYVKVRLDET PAQDEWAHPL VRRQRLNLEQ SLLVAILRQH FVAWEQESGT GASQAQIAID D LLPQLQIY ...String: MAGFFDKLIN RSVTANAGCE PEPSDEEVTD ESVEDSLASS ETRTLQKIRE ATQELLKYGL LEEASKPNLY RIVLSHPEEV TRILEPLDL DIGIDEIRGL LYVKVRLDET PAQDEWAHPL VRRQRLNLEQ SLLVAILRQH FVAWEQESGT GASQAQIAID D LLPQLQIY LGDPGSESKE RTRLLTLLDQ LKGHGLVTSP DAHERIVIRP IIAHLADPIN LQALLAWLRE QIAQQTSPND AP EKDSSEE DVG |
-Macromolecule #3: JetA
| Macromolecule | Name: JetA / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 63.350883 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAHHHHHHHH HHGGSSAWSH PQFEKGGGSG GGSGGGSWSH PQFEKLEVLF QGPAAMEENT RQRTENYISA KNQHPAWILL ATRRAPLVL SCLKTLFEKS HDGIPLEEAI QSLSSILIEH VSQEQYDINQ DNPFLQASRE LREWIKRRLI VERDGRIFAT D ALEVAITF ...String: MAHHHHHHHH HHGGSSAWSH PQFEKGGGSG GGSGGGSWSH PQFEKLEVLF QGPAAMEENT RQRTENYISA KNQHPAWILL ATRRAPLVL SCLKTLFEKS HDGIPLEEAI QSLSSILIEH VSQEQYDINQ DNPFLQASRE LREWIKRRLI VERDGRIFAT D ALEVAITF VESLDNRFMT STASRLSTVQ REIENLETRL NPNPANRVAT LRRRISELER ELQEAEAGHI EVLETHQAVE HI RDVYNLA SSLRADFRRV EDSWREADRA LRQSIIGEQY HRGDIVERLL NDQDALLNTP EGRVFDSFQQ QLRQSSELKA MSE RLRVIL SHPSASDALN RLQRHDLRWL VKRLVDESQT VLQARARSER DVRGFMKTGL AAEHHRVGHL LNEFLNLALK LDWQ RQMIR KQEVPLPAVG VAVTGIPAIE RLRFKEVDDE AEQTLDLSNH AADLTQIGDD FWDAFNGLDR EVLIQQTLQL LAKEN RPVG LAELAELLPP AHDLETFAVW IGMAREAGIE VIDSQREFAE LSDGEGRRWR FNLPTTGLES QALMDIDWEG |
-Macromolecule #4: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)