[English] 日本語
 Yorodumi
Yorodumi- EMDB-15575: Vaccinia virus DNA helicase D5 residues 323-785 hexamer with boun... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Vaccinia virus DNA helicase D5 residues 323-785 hexamer with bound DNA processed in C1 | ||||||||||||||||||
 Map data Map data | Masked map in C1 from Relion PostProcess | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | DNA helicase / D5_N domain / DUF5906 domain / Pox_D5 domain / SF3 helicase / VIRAL PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhelicase activity / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / host cell cytoplasm / hydrolase activity / ATP binding Similarity search - Function | ||||||||||||||||||
| Biological species |  Vaccinia virus Copenhagen / Synthetic construct (others) / synthetic construct (others) Vaccinia virus Copenhagen / Synthetic construct (others) / synthetic construct (others) | ||||||||||||||||||
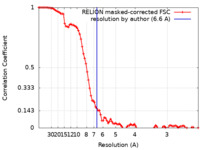
| Method | single particle reconstruction / cryo EM / Resolution: 6.6 Å | ||||||||||||||||||
 Authors Authors | Burmeister WP / Hutin S / Ling WL / Grimm C / Schoehn G | ||||||||||||||||||
| Funding support |  France, 5 items France, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Viruses / Year: 2022 Journal: Viruses / Year: 2022Title: The Vaccinia Virus DNA Helicase Structure from Combined Single-Particle Cryo-Electron Microscopy and AlphaFold2 Prediction. Authors: Stephanie Hutin / Wai Li Ling / Nicolas Tarbouriech / Guy Schoehn / Clemens Grimm / Utz Fischer / Wim P Burmeister /   Abstract: Poxviruses are large DNA viruses with a linear double-stranded DNA genome circularized at the extremities. The helicase-primase D5, composed of six identical 90 kDa subunits, is required for DNA ...Poxviruses are large DNA viruses with a linear double-stranded DNA genome circularized at the extremities. The helicase-primase D5, composed of six identical 90 kDa subunits, is required for DNA replication. D5 consists of a primase fragment flexibly attached to the hexameric C-terminal polypeptide (res. 323-785) with confirmed nucleotide hydrolase and DNA-binding activity but an elusive helicase activity. We determined its structure by single-particle cryo-electron microscopy. It displays an AAA+ helicase core flanked by N- and C-terminal domains. Model building was greatly helped by the predicted structure of D5 using AlphaFold2. The 3.9 Å structure of the N-terminal domain forms a well-defined tight ring while the resolution decreases towards the C-terminus, still allowing the fit of the predicted structure. The N-terminal domain is partially present in papillomavirus E1 and polyomavirus LTA helicases, as well as in a bacteriophage NrS-1 helicase domain, which is also closely related to the AAA+ helicase domain of D5. Using the Pfam domain database, a D5_N domain followed by DUF5906 and Pox_D5 domains could be assigned to the cryo-EM structure, providing the first 3D structures for D5_N and Pox_D5 domains. The same domain organization has been identified in a family of putative helicases from large DNA viruses, bacteriophages, and selfish DNA elements. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15575.map.gz emd_15575.map.gz | 5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15575-v30.xml emd-15575-v30.xml emd-15575.xml emd-15575.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15575_fsc.xml emd_15575_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_15575.png emd_15575.png | 161.2 KB | ||
| Others |  emd_15575_half_map_1.map.gz emd_15575_half_map_1.map.gz emd_15575_half_map_2.map.gz emd_15575_half_map_2.map.gz | 49.6 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15575 http://ftp.pdbj.org/pub/emdb/structures/EMD-15575 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15575 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15575 | HTTPS FTP |
-Validation report
| Summary document |  emd_15575_validation.pdf.gz emd_15575_validation.pdf.gz | 750.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15575_full_validation.pdf.gz emd_15575_full_validation.pdf.gz | 750.4 KB | Display | |
| Data in XML |  emd_15575_validation.xml.gz emd_15575_validation.xml.gz | 16.1 KB | Display | |
| Data in CIF |  emd_15575_validation.cif.gz emd_15575_validation.cif.gz | 21.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15575 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15575 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15575 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15575 | HTTPS FTP |
-Related structure data
| Related structure data |  8apmMC  8aplC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15575.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15575.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked map in C1 from Relion PostProcess | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.21 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map from Relion Refine3D in C1
| File | emd_15575_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map from Relion Refine3D in C1 | ||||||||||||
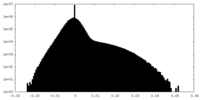
| Projections & Slices |
| ||||||||||||
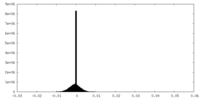
| Density Histograms |
-Half map: Half map from Relion Refine3D in C1
| File | emd_15575_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map from Relion Refine3D in C1 | ||||||||||||
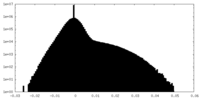
| Projections & Slices |
| ||||||||||||
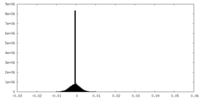
| Density Histograms |
- Sample components
Sample components
-Entire : Primase D5 C-terminal fragment res. 323 - res. 785
| Entire | Name: Primase D5 C-terminal fragment res. 323 - res. 785 |
|---|---|
| Components |
|
-Supramolecule #1: Primase D5 C-terminal fragment res. 323 - res. 785
| Supramolecule | Name: Primase D5 C-terminal fragment res. 323 - res. 785 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: construct |
|---|---|
| Molecular weight | Theoretical: 325 KDa |
-Supramolecule #2: Primase-helicase D5 fragment residues 323-785
| Supramolecule | Name: Primase-helicase D5 fragment residues 323-785 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: Obtained after TEV (tobacco etch virus) cleavage of a construct |
|---|---|
| Source (natural) | Organism:  Vaccinia virus Copenhagen Vaccinia virus Copenhagen |
-Supramolecule #3: DNA (5'-D(P*CP*CP*GP*AP*AP*TP*CP*A)-3')
| Supramolecule | Name: DNA (5'-D(P*CP*CP*GP*AP*AP*TP*CP*A)-3') / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism: Synthetic construct (others) / Synthetically produced: Yes |
-Supramolecule #4: DNA (5'-D(P*TP*GP*AP*TP*TP*CP*GP*G)-3')
| Supramolecule | Name: DNA (5'-D(P*TP*GP*AP*TP*TP*CP*GP*G)-3') / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism: Synthetic construct (others) / Synthetically produced: Yes |
-Macromolecule #1: Primase D5
| Macromolecule | Name: Primase D5 / type: protein_or_peptide / ID: 1 Details: obtained after tobacco etch virus protease cleavage with 2 additional N-terminal residues from the cleavage site. Number of copies: 6 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Vaccinia virus Copenhagen / Strain: Copenhagen Vaccinia virus Copenhagen / Strain: Copenhagen |
| Molecular weight | Theoretical: 53.495285 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AMGNKLFNIA QRILDTNSVL LTERGDYIVW INNSWKFNSE EPLITKLILS IRHQLPKEYS SELLCPRKRK TVEANIRDML VDSVETDTY PDKLPFKNGV LDLVDGMFYS GDDAKKYTCT VSTGFKFDDT KFVEDSPEME ELMNIINDIQ PLTDENKKNR E LYEKTLSS ...String: AMGNKLFNIA QRILDTNSVL LTERGDYIVW INNSWKFNSE EPLITKLILS IRHQLPKEYS SELLCPRKRK TVEANIRDML VDSVETDTY PDKLPFKNGV LDLVDGMFYS GDDAKKYTCT VSTGFKFDDT KFVEDSPEME ELMNIINDIQ PLTDENKKNR E LYEKTLSS CLCGATKGCL TFFFGETATG KSTTKRLLKS AIGDLFVETG QTILTDVLDK GPNPFIANMH LKRSVFCSEL PD FACSGSK KIRSDNIKKL TEPCVIGRPC FSNKINNRNH ATIIIDTNYK PVFDRIDNAL MRRIAVVRFR THFSQPSGRE AAE NNDAYD KVKLLDEGLD GKIQNNRYRF AFLYLLVKWY KKYHVPIMKL YPTPEEIPDF AFYLKIGTLL VSSSVKHIPL MTDL SKKGY ILYDNVVTLP LTTFQQKISK YFNSRLFGHD IESFINRHKK FANVSDEYLQ YIFIEDISSP UniProtKB: Uncoating factor OPG117 |
-Macromolecule #2: DNA (5'-D(P*CP*CP*GP*AP*AP*TP*CP*A)-3')
| Macromolecule | Name: DNA (5'-D(P*CP*CP*GP*AP*AP*TP*CP*A)-3') / type: dna / ID: 2 Details: synthetic, at the current resolution the sequence does not matter, the DNA oligomer was end-labelled: biotin-ccgaatcaggaagataacagcggtttagcc3-digoxigenin Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 9.265994 KDa |
| Sequence | String: (DC)(DC)(DG)(DA)(DA)(DT)(DC)(DA)(DG)(DG) (DA)(DA)(DG)(DA)(DT)(DA)(DA)(DC)(DA)(DG) (DC)(DG)(DG)(DT)(DT)(DT)(DA)(DG)(DC) (DC) |
-Macromolecule #3: DNA (5'-D(P*TP*GP*AP*TP*TP*CP*GP*G)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*GP*AP*TP*TP*CP*GP*G)-3') / type: dna / ID: 3 Details: At the current resolution, the base mismatch does not have any impact as it is located in an unpaired part of the DNA. Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 9.21191 KDa |
| Sequence | String: (DG)(DG)(DC)(DT)(DT)(DA)(DG)(DT)(DC)(DC) (DT)(DT)(DC)(DT)(DA)(DT)(DT)(DG)(DT)(DC) (DG)(DC)(DA)(DG)(DA)(DT)(DT)(DC)(DG) (DG) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.24 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| |||||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||||||||
| Details | The sample contained also a dsDNA oligomer in slight stoechiometric excess over hexamers. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 830 / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 152 |
|---|---|
| Output model |  PDB-8apm: |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X