+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tube assembly of Atg18-PR72AA | |||||||||
 Map data Map data | sharpened map from CryoSPARC software | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | autophagy / membrane remodeling / PIP binding / PI3P / PI(3 / 5)P2 / lipid binding protein / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of phosphatidylinositol biosynthetic process / PAS complex / 1-phosphatidyl-1D-myo-inositol 3,5-bisphosphate metabolic process / phagophore / positive regulation of vacuole organization / vacuolar protein processing / Macroautophagy / glycophagy / autophagy of peroxisome / cytoplasm to vacuole targeting by the Cvt pathway ...regulation of phosphatidylinositol biosynthetic process / PAS complex / 1-phosphatidyl-1D-myo-inositol 3,5-bisphosphate metabolic process / phagophore / positive regulation of vacuole organization / vacuolar protein processing / Macroautophagy / glycophagy / autophagy of peroxisome / cytoplasm to vacuole targeting by the Cvt pathway / nucleophagy / pexophagy / protein localization to phagophore assembly site / phagophore assembly site membrane / late endosome to vacuole transport / piecemeal microautophagy of the nucleus / phosphatidylinositol-3-phosphate binding / fungal-type vacuole membrane / phagophore assembly site / phosphatidylinositol-4-phosphate binding / phosphatidylinositol-3,5-bisphosphate binding / vacuolar membrane / extrinsic component of membrane / autophagy of mitochondrion / autophagosome assembly / ubiquitin binding / cell periphery / macroautophagy / endosome membrane / endosome / protein-containing complex / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
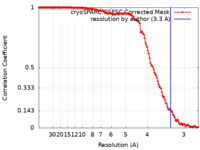
| Method | helical reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Mann D / Fromm S / Martinez-Sanchez A / Gopaldass N / Mayer A / Sachse C | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds. Authors: Daniel Mann / Simon A Fromm / Antonio Martinez-Sanchez / Navin Gopaldass / Ramona Choy / Andreas Mayer / Carsten Sachse /    Abstract: Autophagy-related protein 18 (Atg18) participates in the elongation of early autophagosomal structures in concert with Atg2 and Atg9 complexes. How Atg18 contributes to the structural coordination of ...Autophagy-related protein 18 (Atg18) participates in the elongation of early autophagosomal structures in concert with Atg2 and Atg9 complexes. How Atg18 contributes to the structural coordination of Atg2 and Atg9 at the isolation membrane remains to be understood. Here, we determined the cryo-EM structures of Atg18 organized in helical tubes, Atg18 oligomers in solution as well as on lipid membrane scaffolds. The helical assembly is composed of Atg18 tetramers forming a lozenge cylindrical lattice with remarkable structural similarity to the COPII outer coat. When reconstituted with lipid membranes, using subtomogram averaging we determined tilted Atg18 dimer structures bridging two juxtaposed lipid membranes spaced apart by 80 Å. Moreover, lipid reconstitution experiments further delineate the contributions of Atg18's FRRG motif and the amphipathic helical extension in membrane interaction. The observed structural plasticity of Atg18's oligomeric organization and membrane binding properties provide a molecular framework for the positioning of downstream components of the autophagy machinery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15408.map.gz emd_15408.map.gz | 229.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15408-v30.xml emd-15408-v30.xml emd-15408.xml emd-15408.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15408_fsc.xml emd_15408_fsc.xml | 13.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_15408.png emd_15408.png | 284.1 KB | ||
| Masks |  emd_15408_msk_1.map emd_15408_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15408.cif.gz emd-15408.cif.gz | 5.7 KB | ||
| Others |  emd_15408_additional_1.map.gz emd_15408_additional_1.map.gz emd_15408_half_map_1.map.gz emd_15408_half_map_1.map.gz emd_15408_half_map_2.map.gz emd_15408_half_map_2.map.gz | 186.4 MB 226.2 MB 226.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15408 http://ftp.pdbj.org/pub/emdb/structures/EMD-15408 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15408 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15408 | HTTPS FTP |
-Validation report
| Summary document |  emd_15408_validation.pdf.gz emd_15408_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15408_full_validation.pdf.gz emd_15408_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_15408_validation.xml.gz emd_15408_validation.xml.gz | 21.5 KB | Display | |
| Data in CIF |  emd_15408_validation.cif.gz emd_15408_validation.cif.gz | 28 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15408 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15408 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15408 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15408 | HTTPS FTP |
-Related structure data
| Related structure data |  8afqMC  8afwC  8afxC  8afyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15408.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15408.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map from CryoSPARC software | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15408_msk_1.map emd_15408_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: autosharpened map from Phenix software
| File | emd_15408_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | autosharpened map from Phenix software | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_15408_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_15408_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Filament assembly of Atg18-PR72AA
| Entire | Name: Filament assembly of Atg18-PR72AA |
|---|---|
| Components |
|
-Supramolecule #1: Filament assembly of Atg18-PR72AA
| Supramolecule | Name: Filament assembly of Atg18-PR72AA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Autophagy-related protein 18
| Macromolecule | Name: Autophagy-related protein 18 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 55.045777 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSDSSPTINF INFNQTGTCI SLGTSKGFKI FNCEPFGKFY SEDSGGYAIV EMLFSTSLLA LVGIGDQPAL SAARLRIINT KKHSIICEV TFPTSILSVK MNKSRLVVLL QEQIYIYDIN TMRLLHTIET NPNPRGLMAM SPSVANSYLV YPSPPKVINS E IKAHATTN ...String: MSDSSPTINF INFNQTGTCI SLGTSKGFKI FNCEPFGKFY SEDSGGYAIV EMLFSTSLLA LVGIGDQPAL SAARLRIINT KKHSIICEV TFPTSILSVK MNKSRLVVLL QEQIYIYDIN TMRLLHTIET NPNPRGLMAM SPSVANSYLV YPSPPKVINS E IKAHATTN NITLSVGGNT ETSFKRDQQD AGHSDISDLD QYSSFTKRDD ADPTSSNGGN SSIIKNGDVI VFNLETLQPT MV IEAHKGE IAAMAISFDG TLMATASDKG TIIRVFDIET GDKIYQFRRG TYATRIYSIS FSEDSQYLAV TGSSKTVHIF KLG HSMSNN KLDSDDSNME EAAADDSSLD TTSIDALSDE ENPTRLAREP YVDASRKTMG RMIRYSSQKL SRRAARTLGQ IFPI KVTSL LESSRHFASL KLPVETNSHV MTISSIGSPI DIDTSEYPEL FETGNSASTE SYHEPVMKMV PIRVVSSDGY LYNFV MDPE RGGDCLILSQ YSILMD UniProtKB: Autophagy-related protein 18 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 99 % / Chamber temperature: 291 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 2380 / Average electron dose: 90.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8afq: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)