[English] 日本語
 Yorodumi
Yorodumi- EMDB-15256: Composite map of the C. reinhardtii ciliary transition zone (stru... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Composite map of the C. reinhardtii ciliary transition zone (structures attached to a single MTD) from in situ subtomogram averaging | ||||||||||||||||||||||||
 Map data Map data | Composite map of central TZ structures (Stellate, Y-link) | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | cilia / transition zone / flagella / ciliary pore / intraflagellar transport / motility / signaling / basal body / microtubules / chlamydomonas / cytoskeleton / MTD / doublet / stellate / Y-link / STRUCTURAL PROTEIN | ||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 28.2 Å | ||||||||||||||||||||||||
 Authors Authors | van den Hoek HG / Righetto RD / Schaffer M / Erdmann PS / Wan WN / Plitzko JM / Baumeister W / Engel BD | ||||||||||||||||||||||||
| Funding support | European Union,  Germany, Germany,  Switzerland, 7 items Switzerland, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: In situ architecture of the ciliary base reveals the stepwise assembly of intraflagellar transport trains. Authors: Hugo van den Hoek / Nikolai Klena / Mareike A Jordan / Gonzalo Alvarez Viar / Ricardo D Righetto / Miroslava Schaffer / Philipp S Erdmann / William Wan / Stefan Geimer / Jürgen M Plitzko / ...Authors: Hugo van den Hoek / Nikolai Klena / Mareike A Jordan / Gonzalo Alvarez Viar / Ricardo D Righetto / Miroslava Schaffer / Philipp S Erdmann / William Wan / Stefan Geimer / Jürgen M Plitzko / Wolfgang Baumeister / Gaia Pigino / Virginie Hamel / Paul Guichard / Benjamin D Engel /     Abstract: The cilium is an antenna-like organelle that performs numerous cellular functions, including motility, sensing, and signaling. The base of the cilium contains a selective barrier that regulates the ...The cilium is an antenna-like organelle that performs numerous cellular functions, including motility, sensing, and signaling. The base of the cilium contains a selective barrier that regulates the entry of large intraflagellar transport (IFT) trains, which carry cargo proteins required for ciliary assembly and maintenance. However, the native architecture of the ciliary base and the process of IFT train assembly remain unresolved. In this work, we used in situ cryo-electron tomography to reveal native structures of the transition zone region and assembling IFT trains at the ciliary base in . We combined this direct cellular visualization with ultrastructure expansion microscopy to describe the front-to-back stepwise assembly of IFT trains: IFT-B forms the backbone, onto which bind IFT-A, dynein-1b, and finally kinesin-2 before entry into the cilium. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15256.map.gz emd_15256.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15256-v30.xml emd-15256-v30.xml emd-15256.xml emd-15256.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15256.png emd_15256.png | 70.5 KB | ||
| Filedesc metadata |  emd-15256.cif.gz emd-15256.cif.gz | 4.8 KB | ||
| Others |  emd_15256_additional_1.map.gz emd_15256_additional_1.map.gz emd_15256_additional_2.map.gz emd_15256_additional_2.map.gz | 2.4 MB 925.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15256 http://ftp.pdbj.org/pub/emdb/structures/EMD-15256 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15256 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15256 | HTTPS FTP |
-Validation report
| Summary document |  emd_15256_validation.pdf.gz emd_15256_validation.pdf.gz | 321.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15256_full_validation.pdf.gz emd_15256_full_validation.pdf.gz | 321.4 KB | Display | |
| Data in XML |  emd_15256_validation.xml.gz emd_15256_validation.xml.gz | 6.2 KB | Display | |
| Data in CIF |  emd_15256_validation.cif.gz emd_15256_validation.cif.gz | 7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15256 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15256 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15256 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15256 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15256.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15256.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map of central TZ structures (Stellate, Y-link) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.84 Å | ||||||||||||||||||||||||||||||||||||
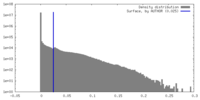
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Composite map: Stellate component
| File | emd_15256_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map: Stellate component | ||||||||||||
| Projections & Slices |
| ||||||||||||
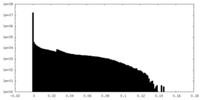
| Density Histograms |
-Additional map: Composite map: Y-link component
| File | emd_15256_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map: Y-link component | ||||||||||||
| Projections & Slices |
| ||||||||||||
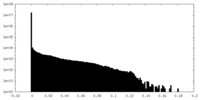
| Density Histograms |
- Sample components
Sample components
-Entire : Composite map of MTD-bound ciliary transition zone structures in ...
| Entire | Name: Composite map of MTD-bound ciliary transition zone structures in C. reinhardtii |
|---|---|
| Components |
|
-Supramolecule #1: Composite map of MTD-bound ciliary transition zone structures in ...
| Supramolecule | Name: Composite map of MTD-bound ciliary transition zone structures in C. reinhardtii type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 100.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 4.0 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 28.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: STOPGAP (ver. 0.3) Details: Composite map generated from two separate subtomogram averages Stellate: 206 particles, 28.2 A Y-link: 529 particles, 27.9 A Number subtomograms used: 206 |
|---|---|
| Extraction | Number tomograms: 19 / Number images used: 1210 / Software - Name:  IMOD IMOD |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: STOPGAP (ver. 0.3) |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)