[English] 日本語
 Yorodumi
Yorodumi- EMDB-15162: Structure of the human mitochondrial HSPD1 single ring from grids... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human mitochondrial HSPD1 single ring from grids produced in 14ms | |||||||||
 Map data Map data | Final postprocessed map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
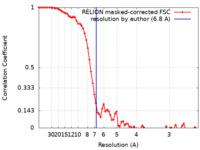
| Method | single particle reconstruction / cryo EM / Resolution: 6.8 Å | |||||||||
 Authors Authors | Klebl DP / Wang Y / Thompson RF / Muench SP | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Front Mol Biosci / Year: 2022 Journal: Front Mol Biosci / Year: 2022Title: It started with a Cys: Spontaneous cysteine modification during cryo-EM grid preparation. Authors: David P Klebl / Yiheng Wang / Frank Sobott / Rebecca F Thompson / Stephen P Muench /  Abstract: Advances in single particle cryo-EM data collection and processing have seen a significant rise in its use. However, the influences of the environment generated through grid preparation, by for ...Advances in single particle cryo-EM data collection and processing have seen a significant rise in its use. However, the influences of the environment generated through grid preparation, by for example interactions of proteins with the air-water interface are poorly understood and can be a major hurdle in structure determination by cryo-EM. Initial interactions of proteins with the air-water interface occur quickly and proteins can adopt preferred orientation or partially unfold within hundreds of milliseconds. It has also been shown previously that thin-film layers create hydroxyl radicals. To investigate the potential this might have in cryo-EM sample preparation, we studied two proteins, HSPD1, and beta-galactosidase, and show that cysteine residues are modified in a time-dependent manner. In the case of both HSPD1 and beta-galactosidase, this putative oxidation is linked to partial protein unfolding, as well as more subtle structural changes. We show these modifications can be alleviated through increasing the speed of grid preparation, the addition of DTT, or by sequestering away from the AWI using continuous support films. We speculate that the modification is oxidation by reactive oxygen species which are formed and act at the air-water interface. Finally, we show grid preparation on a millisecond timescale outruns cysteine modification, showing that the reaction timescale is in the range of 100s to 1,000s milliseconds and offering an alternative approach to prevent spontaneous cysteine modification and its consequences during cryo-EM grid preparation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15162.map.gz emd_15162.map.gz | 4.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15162-v30.xml emd-15162-v30.xml emd-15162.xml emd-15162.xml | 13.2 KB 13.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15162_fsc.xml emd_15162_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_15162.png emd_15162.png | 49.1 KB | ||
| Masks |  emd_15162_msk_1.map emd_15162_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Others |  emd_15162_half_map_1.map.gz emd_15162_half_map_1.map.gz emd_15162_half_map_2.map.gz emd_15162_half_map_2.map.gz | 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15162 http://ftp.pdbj.org/pub/emdb/structures/EMD-15162 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15162 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15162 | HTTPS FTP |
-Validation report
| Summary document |  emd_15162_validation.pdf.gz emd_15162_validation.pdf.gz | 709.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15162_full_validation.pdf.gz emd_15162_full_validation.pdf.gz | 708.9 KB | Display | |
| Data in XML |  emd_15162_validation.xml.gz emd_15162_validation.xml.gz | 12.8 KB | Display | |
| Data in CIF |  emd_15162_validation.cif.gz emd_15162_validation.cif.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15162 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15162 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15162 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15162 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15162.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15162.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final postprocessed map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.23 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15162_msk_1.map emd_15162_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: second Half map for fast HSPD1
| File | emd_15162_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | second Half map for fast HSPD1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map for fast HSPD1
| File | emd_15162_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map for fast HSPD1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : human mitochondrial heat shock protein family member D1 (HSPD1)
| Entire | Name: human mitochondrial heat shock protein family member D1 (HSPD1) |
|---|---|
| Components |
|
-Supramolecule #1: human mitochondrial heat shock protein family member D1 (HSPD1)
| Supramolecule | Name: human mitochondrial heat shock protein family member D1 (HSPD1) type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Experimental: 60 kDa/nm |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.3 mg/mL |
|---|---|
| Buffer | pH: 7.7 / Details: 25 mM Tris, 150 mM NaCl, 5 mM MgCl2 pH 7.7 |
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: HOMEMADE PLUNGER / Details: Home-made high speed plunging device. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 56.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.3000000000000003 µm / Nominal defocus min: 1.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)