[English] 日本語
 Yorodumi
Yorodumi- EMDB-15119: 13pf undecorated microtubule from recombinant human tubulin (alph... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 13pf undecorated microtubule from recombinant human tubulin (alpha1B, beta3) with spliced unmodified C-terminal tail on alpha1B. | |||||||||
 Map data Map data | Human microtubules (alpha1B, beta3) with spliced unmodified C-terminal tail on alpha1B. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | tubulin / no tail / recombinant / semisynthetic / STRUCTURAL PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
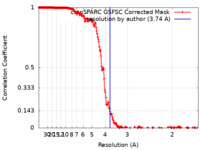
| Method | helical reconstruction / cryo EM / Resolution: 3.74 Å | |||||||||
 Authors Authors | Ebberink E / Fernandes S / Hatzopoulos GN / Agashe N / Guidotti N / Reichart T / Reymond L / Velluz MC / Schneider FZ / Pourroy C ...Ebberink E / Fernandes S / Hatzopoulos GN / Agashe N / Guidotti N / Reichart T / Reymond L / Velluz MC / Schneider FZ / Pourroy C / Janke C / Gonczy P / Aumeier C / Fierz B | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem / Year: 2023 Journal: Nat Chem / Year: 2023Title: Tubulin engineering by semi-synthesis reveals that polyglutamylation directs detyrosination. Authors: Eduard Ebberink / Simon Fernandes / Georgios Hatzopoulos / Ninad Agashe / Po-Han Chang / Nora Guidotti / Timothy M Reichart / Luc Reymond / Marie-Claire Velluz / Fabian Schneider / Cédric ...Authors: Eduard Ebberink / Simon Fernandes / Georgios Hatzopoulos / Ninad Agashe / Po-Han Chang / Nora Guidotti / Timothy M Reichart / Luc Reymond / Marie-Claire Velluz / Fabian Schneider / Cédric Pourroy / Carsten Janke / Pierre Gönczy / Beat Fierz / Charlotte Aumeier /   Abstract: Microtubules, a critical component of the cytoskeleton, carry post-translational modifications (PTMs) that are important for the regulation of key cellular processes. Long-lived microtubules, in ...Microtubules, a critical component of the cytoskeleton, carry post-translational modifications (PTMs) that are important for the regulation of key cellular processes. Long-lived microtubules, in neurons particularly, exhibit both detyrosination of α-tubulin and polyglutamylation. Dysregulation of these PTMs can result in developmental defects and neurodegeneration. Owing to a lack of tools to study the regulation and function of these PTMs, the mechanisms that govern such PTM patterns are not well understood. Here we produce fully functional tubulin carrying precisely defined PTMs within its C-terminal tail. We ligate synthetic α-tubulin tails-which are site-specifically glutamylated-to recombinant human tubulin heterodimers by applying a sortase- and intein-mediated tandem transamidation strategy. Using microtubules reconstituted with these designer tubulins, we find that α-tubulin polyglutamylation promotes its detyrosination by enhancing the activity of the tubulin tyrosine carboxypeptidase vasohibin/small vasohibin-binding protein in a manner dependent on the length of polyglutamyl chains. We also find that modulating polyglutamylation levels in cells results in corresponding changes in detyrosination, corroborating the link between the detyrosination cycle to polyglutamylation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15119.map.gz emd_15119.map.gz | 767.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15119-v30.xml emd-15119-v30.xml emd-15119.xml emd-15119.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15119_fsc.xml emd_15119_fsc.xml | 20 KB | Display |  FSC data file FSC data file |
| Images |  emd_15119.png emd_15119.png | 126.3 KB | ||
| Masks |  emd_15119_msk_1.map emd_15119_msk_1.map | 824 MB |  Mask map Mask map | |
| Others |  emd_15119_half_map_1.map.gz emd_15119_half_map_1.map.gz emd_15119_half_map_2.map.gz emd_15119_half_map_2.map.gz | 763.4 MB 763.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15119 http://ftp.pdbj.org/pub/emdb/structures/EMD-15119 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15119 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15119 | HTTPS FTP |
-Validation report
| Summary document |  emd_15119_validation.pdf.gz emd_15119_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15119_full_validation.pdf.gz emd_15119_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_15119_validation.xml.gz emd_15119_validation.xml.gz | 28 KB | Display | |
| Data in CIF |  emd_15119_validation.cif.gz emd_15119_validation.cif.gz | 36.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15119 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15119 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15119 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15119 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15119.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15119.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human microtubules (alpha1B, beta3) with spliced unmodified C-terminal tail on alpha1B. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
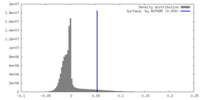
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15119_msk_1.map emd_15119_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
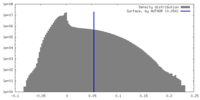
| Density Histograms |
-Half map: #1
| File | emd_15119_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
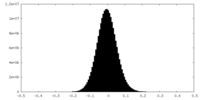
| Density Histograms |
-Half map: #2
| File | emd_15119_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
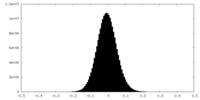
| Density Histograms |
- Sample components
Sample components
-Entire : 13pf microtubule from recombinant human tubulin with spliced unmo...
| Entire | Name: 13pf microtubule from recombinant human tubulin with spliced unmodified C-terminal tail on alpha1B. |
|---|---|
| Components |
|
-Supramolecule #1: 13pf microtubule from recombinant human tubulin with spliced unmo...
| Supramolecule | Name: 13pf microtubule from recombinant human tubulin with spliced unmodified C-terminal tail on alpha1B. type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Tubulin alpha-1B with spliced C-terminal tail
| Macromolecule | Name: Tubulin alpha-1B with spliced C-terminal tail / type: protein_or_peptide / ID: 1 Details: internal 6xHis tag in 40-loop; Spliced C-terminal tail without glutamylation, splicing scar: InsC440 and V441F Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MRECISIHVG QAGVQIGNA C WELYCLEH GI QPDGQMP SDK TIHHHH HHGGGDDSFN TFFSETGAG K HVPRAVFV DL EPTVIDE VRT GTYRQL FHPE QLITG KEDAA NNYA RGHYTI GKE IIDLVLD RI RKLADQCT G LQGFLVFHS FGGGTGSGFT ...String: MRECISIHVG QAGVQIGNA C WELYCLEH GI QPDGQMP SDK TIHHHH HHGGGDDSFN TFFSETGAG K HVPRAVFV DL EPTVIDE VRT GTYRQL FHPE QLITG KEDAA NNYA RGHYTI GKE IIDLVLD RI RKLADQCT G LQGFLVFHS FGGGTGSGFT SLLMERLSV D YGKKSKLE FS IYPAPQV STA VVEPYN SILT THTTL EHSDC AFMV DNEAIY DIC RRNLDIE RP TYTNLNRL I SQIVSSITA SLRFDGALNV DLTEFQTNL V PYPRIHFP LA TYAPVIS AEK AYHEQL SVAE ITNAC FEPAN QMVK CDPRHG KYM ACCLLYR GD VVPKDVNA A IATIKTKRS IQFVDWCPTG FKVGINYQP P TVVPGGDL AK VQRAVCM LSN TTAIAE AWAR LDHKF DLMYA KRAF VHWYVG EGM EEGEFSE AR EDMAALEK D YEEVGVDSC FEGEGEEEGE EY |
-Macromolecule #2: Tubulin beta-3
| Macromolecule | Name: Tubulin beta-3 / type: protein_or_peptide / ID: 2 / Details: C-terminal FLAG-tag / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPSGNYVGDS DLQLERISVY YNEASSHKYV PRAILVDLEP GTMDSVRSGA FGHLFRPDNF IFGQSGAGNN WAKGHYTEGA ELVDSVLDVV RKECENCDCL QGFQLTHSLG GGTGSGMGTL LISKVREEYP DRIMNTFSVV ...String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPSGNYVGDS DLQLERISVY YNEASSHKYV PRAILVDLEP GTMDSVRSGA FGHLFRPDNF IFGQSGAGNN WAKGHYTEGA ELVDSVLDVV RKECENCDCL QGFQLTHSLG GGTGSGMGTL LISKVREEYP DRIMNTFSVV PSPKVSDTVV EPYNATLSIH QLVENTDETY CIDNEALYDI CFRTLKLATP TYGDLNHLVS ATMSGVTTSL RFPGQLNADL RKLAVNMVPF PRLHFFMPGF APLTARGSQQ YRALTVPELT QQMFDAKNMM AACDPRHGRY LTVATVFRGR MSMKEVDEQM LAIQSKNSSY FVEWIPNNVK VAVCDIPPRG LKMSSTFIGN STAIQELFKR ISEQFTAMFR RKAFLHWYTG EGMDEMEFTE AESNMNDLVS EYQQYQDATA EEEGEMYEDD EEESEAQGPK ENLYFQSSGG DYKDDDDK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)