+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ cryo-electron tomogram of a T. kivui cell 2 | ||||||||||||||||||
 Map data Map data | Cryo-electron tomogram of a T. kivui cell showing HDCR filament bundles. Tomogram was denoised using CRYO-CARE. | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |   Thermoanaerobacter kivui (bacteria) Thermoanaerobacter kivui (bacteria) | ||||||||||||||||||
| Method | electron tomography / cryo EM | ||||||||||||||||||
 Authors Authors | Dietrich HM / Righetto RD / Kumar A / Wietrzynski W / Schuller SK / Trischler R / Wagner J / Schwarz FM / Engel BD / Mueller V / Schuller JM | ||||||||||||||||||
| Funding support |  Germany, European Union, 5 items Germany, European Union, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Membrane-anchored HDCR nanowires drive hydrogen-powered CO fixation. Authors: Helge M Dietrich / Ricardo D Righetto / Anuj Kumar / Wojciech Wietrzynski / Raphael Trischler / Sandra K Schuller / Jonathan Wagner / Fabian M Schwarz / Benjamin D Engel / Volker Müller / Jan M Schuller /   Abstract: Filamentous enzymes have been found in all domains of life, but the advantage of filamentation is often elusive. Some anaerobic, autotrophic bacteria have an unusual filamentous enzyme for CO ...Filamentous enzymes have been found in all domains of life, but the advantage of filamentation is often elusive. Some anaerobic, autotrophic bacteria have an unusual filamentous enzyme for CO fixation-hydrogen-dependent CO reductase (HDCR)-which directly converts H and CO into formic acid. HDCR reduces CO with a higher activity than any other known biological or chemical catalyst, and it has therefore gained considerable interest in two areas of global relevance: hydrogen storage and combating climate change by capturing atmospheric CO. However, the mechanistic basis of the high catalytic turnover rate of HDCR has remained unknown. Here we use cryo-electron microscopy to reveal the structure of a short HDCR filament from the acetogenic bacterium Thermoanaerobacter kivui. The minimum repeating unit is a hexamer that consists of a formate dehydrogenase (FdhF) and two hydrogenases (HydA2) bound around a central core of hydrogenase Fe-S subunits, one HycB3 and two HycB4. These small bacterial polyferredoxin-like proteins oligomerize through their C-terminal helices to form the backbone of the filament. By combining structure-directed mutagenesis with enzymatic analysis, we show that filamentation and rapid electron transfer through the filament enhance the activity of HDCR. To investigate the structure of HDCR in situ, we imaged T. kivui cells with cryo-electron tomography and found that HDCR filaments bundle into large ring-shaped superstructures attached to the plasma membrane. This supramolecular organization may further enhance the stability and connectivity of HDCR to form a specialized metabolic subcompartment within the cell. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15056.map.gz emd_15056.map.gz | 1.4 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15056-v30.xml emd-15056-v30.xml emd-15056.xml emd-15056.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15056.png emd_15056.png | 317.8 KB | ||
| Others |  emd_15056_additional_1.map.gz emd_15056_additional_1.map.gz | 1.4 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15056 http://ftp.pdbj.org/pub/emdb/structures/EMD-15056 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15056 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15056 | HTTPS FTP |
-Validation report
| Summary document |  emd_15056_validation.pdf.gz emd_15056_validation.pdf.gz | 451.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15056_full_validation.pdf.gz emd_15056_full_validation.pdf.gz | 451.1 KB | Display | |
| Data in XML |  emd_15056_validation.xml.gz emd_15056_validation.xml.gz | 4.6 KB | Display | |
| Data in CIF |  emd_15056_validation.cif.gz emd_15056_validation.cif.gz | 5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15056 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15056 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15056 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15056 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15056.map.gz / Format: CCP4 / Size: 1.5 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15056.map.gz / Format: CCP4 / Size: 1.5 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-electron tomogram of a T. kivui cell showing HDCR filament bundles. Tomogram was denoised using CRYO-CARE. | ||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 14.08 Å | ||||||||||||||||||||||||||||||||
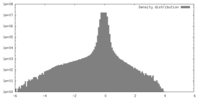
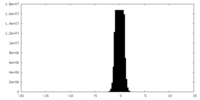
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Cryo-electron tomogram of a T. kivui cell showing...
| File | emd_15056_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-electron tomogram of a T. kivui cell showing HDCR filament bundles. Original reconstruction from IMOD. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : T. kivui cell
| Entire | Name: T. kivui cell |
|---|---|
| Components |
|
-Supramolecule #1: T. kivui cell
| Supramolecule | Name: T. kivui cell / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Thermoanaerobacter kivui (bacteria) Thermoanaerobacter kivui (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: FEI VITROBOT MARK IV |
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 30 kV / Focused ion beam - Current: 0.03 nA / Focused ion beam - Duration: 1800 sec. / Focused ion beam - Temperature: 91 K / Focused ion beam - Initial thickness: 2000 nm / Focused ion beam - Final thickness: 150 nm Focused ion beam - Details: See https://bio-protocol.org/e1575 for detailed procedure.. The value given for _emd_sectioning_focused_ion_beam.instrument is FEI Aquilos FIB. This is not in a list of ...Focused ion beam - Details: See https://bio-protocol.org/e1575 for detailed procedure.. The value given for _emd_sectioning_focused_ion_beam.instrument is FEI Aquilos FIB. This is not in a list of allowed values {'DB235', 'OTHER'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Details | Dose-symmetric tilt-series were acquired (Hagen et al., 2017), starting at +10 degrees to match the pre-tilt of the lamella (i.e. from -50 to +70 degrees). |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.5 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Software - Name:  IMOD / Number images used: 60 IMOD / Number images used: 60 |
|---|---|
| CTF correction | Software: (Name: CTFFIND (ver. 4),  IMOD, NOVACTF) IMOD, NOVACTF) |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)