+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Influenza A/H7N9 polymerase apo-protein dimer complex | |||||||||
 Map data Map data | LocScale filtered map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Influenza / H7N9 / viral RNA-dependent RNA polymerase / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcap snatching / viral transcription / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / virion component / endonuclease activity / host cell cytoplasm / Hydrolases; Acting on ester bonds / RNA-directed RNA polymerase ...cap snatching / viral transcription / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / virion component / endonuclease activity / host cell cytoplasm / Hydrolases; Acting on ester bonds / RNA-directed RNA polymerase / viral translational frameshifting / viral RNA genome replication / RNA-dependent RNA polymerase activity / nucleotide binding / DNA-templated transcription / host cell nucleus / RNA binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Influenza A virus (A/Zhejiang/DTID-ZJU01/2013(H7N9)) Influenza A virus (A/Zhejiang/DTID-ZJU01/2013(H7N9)) | |||||||||
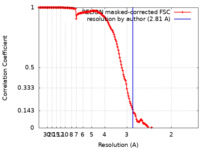
| Method | single particle reconstruction / cryo EM / Resolution: 2.81 Å | |||||||||
 Authors Authors | Cusack S / Kouba T | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2023 Journal: Cell Rep / Year: 2023Title: Direct observation of backtracking by influenza A and B polymerases upon consecutive incorporation of the nucleoside analog T1106. Authors: Tomas Kouba / Anna Dubankova / Petra Drncova / Elisa Donati / Pietro Vidossich / Valentina Speranzini / Alex Pflug / Johanna Huchting / Chris Meier / Marco De Vivo / Stephen Cusack /    Abstract: The antiviral pseudo-base T705 and its de-fluoro analog T1106 mimic adenine or guanine and can be competitively incorporated into nascent RNA by viral RNA-dependent RNA polymerases. Although ...The antiviral pseudo-base T705 and its de-fluoro analog T1106 mimic adenine or guanine and can be competitively incorporated into nascent RNA by viral RNA-dependent RNA polymerases. Although dispersed, single pseudo-base incorporation is mutagenic, consecutive incorporation causes polymerase stalling and chain termination. Using a template encoding single and then consecutive T1106 incorporation four nucleotides later, we obtained a cryogenic electron microscopy structure of stalled influenza A/H7N9 polymerase. This shows that the entire product-template duplex backtracks by 5 nt, bringing the singly incorporated T1106 to the +1 position, where it forms an unexpected T1106:U wobble base pair. Similar structures show that influenza B polymerase also backtracks after consecutive T1106 incorporation, regardless of whether prior single incorporation has occurred. These results give insight into the unusual mechanism of chain termination by pyrazinecarboxamide base analogs. Consecutive incorporation destabilizes the proximal end of the product-template duplex, promoting irreversible backtracking to a more energetically favorable overall configuration. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14858.map.gz emd_14858.map.gz | 159.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14858-v30.xml emd-14858-v30.xml emd-14858.xml emd-14858.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14858_fsc.xml emd_14858_fsc.xml | 15.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_14858.png emd_14858.png | 75.7 KB | ||
| Filedesc metadata |  emd-14858.cif.gz emd-14858.cif.gz | 6.9 KB | ||
| Others |  emd_14858_additional_1.map.gz emd_14858_additional_1.map.gz emd_14858_half_map_1.map.gz emd_14858_half_map_1.map.gz emd_14858_half_map_2.map.gz emd_14858_half_map_2.map.gz | 304.1 MB 257.3 MB 257.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14858 http://ftp.pdbj.org/pub/emdb/structures/EMD-14858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14858 | HTTPS FTP |
-Validation report
| Summary document |  emd_14858_validation.pdf.gz emd_14858_validation.pdf.gz | 665.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14858_full_validation.pdf.gz emd_14858_full_validation.pdf.gz | 665.3 KB | Display | |
| Data in XML |  emd_14858_validation.xml.gz emd_14858_validation.xml.gz | 23.4 KB | Display | |
| Data in CIF |  emd_14858_validation.cif.gz emd_14858_validation.cif.gz | 30.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14858 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14858 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14858 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14858 | HTTPS FTP |
-Related structure data
| Related structure data |  7zpmMC  7qtlC  7r0eC  7r1fC  7zplC  8bdrC  8be0C  8bekC  8bf5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14858.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14858.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LocScale filtered map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8311 Å | ||||||||||||||||||||||||||||||||||||
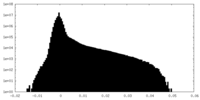
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Relion post-processed map
| File | emd_14858_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion post-processed map | ||||||||||||
| Projections & Slices |
| ||||||||||||
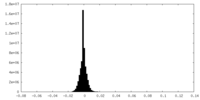
| Density Histograms |
-Half map: #2
| File | emd_14858_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
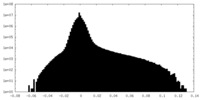
| Density Histograms |
-Half map: #1
| File | emd_14858_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
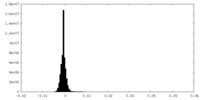
| Density Histograms |
- Sample components
Sample components
-Entire : Influenza A/H7N9 polymerase apo-protein dimer complex
| Entire | Name: Influenza A/H7N9 polymerase apo-protein dimer complex |
|---|---|
| Components |
|
-Supramolecule #1: Influenza A/H7N9 polymerase apo-protein dimer complex
| Supramolecule | Name: Influenza A/H7N9 polymerase apo-protein dimer complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Zhejiang/DTID-ZJU01/2013(H7N9)) Influenza A virus (A/Zhejiang/DTID-ZJU01/2013(H7N9)) |
| Molecular weight | Theoretical: 510 KDa |
-Macromolecule #1: Polymerase acidic protein
| Macromolecule | Name: Polymerase acidic protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Zhejiang/DTID-ZJU01/2013(H7N9)) Influenza A virus (A/Zhejiang/DTID-ZJU01/2013(H7N9))Strain: A/Zhejiang/DTID-ZJU01/2013(H7N9) |
| Molecular weight | Theoretical: 82.930602 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GMEDFVRQCF NPMIVELAEK AMKEYGEDPK IETNKFASIC THLEVCFMYS DFHFIDERGE STIIESGDPN VLLKHRFEII EGRDRTMAW TVVNSICNTT GVEKPKFLPD LYDYKENRFI EIGVTRREVH IYYLEKANKI KSEKTHIHIF SFTGEEMATK A DYTLDEES ...String: GMEDFVRQCF NPMIVELAEK AMKEYGEDPK IETNKFASIC THLEVCFMYS DFHFIDERGE STIIESGDPN VLLKHRFEII EGRDRTMAW TVVNSICNTT GVEKPKFLPD LYDYKENRFI EIGVTRREVH IYYLEKANKI KSEKTHIHIF SFTGEEMATK A DYTLDEES RARIKTRLFT IRQEMASRGL WDSFRQSERG EETIEERFEI TGTMRRLADQ SLPPNFSSLE NFRAYVDGFE PN GCIEGKL SQMSKEVNAR IEPFLRTTPR PLRLPDGPPC SQRSKFLLMD ALKLSIEDPS HEGEGIPLYD AIKCMKTFFG WKE PNIIKP HEKGINPNYL LTWKQVLAEL QDIENEEKIP RTKNMKKTSQ LKWALGENMA PEKVDFEDCK DVNDLKQYDS DEPE PRSLA CWIQSEFNKA CELTDSSWVE LDEIGEDVAP IEHIASMRRN YFTAEVSHCR ATEYIMKGVY INTALLNASC AAMDD FQLI PMISKCRTKE GRRKTNLYGF IIKGRSHLRN DTDVVNFVSM EFSLTDPRLE PHKWEKYCVL EIGDMLLRTA VGQVSR PMF LYVRTNGTSK IKMKWGMEMR RCLLQSLQQI ESMIEAESSV KEKDLTKEFF ENKSETWPIG ESPKGVEEGS IGKVCRT LL AKSVFNSLYA SPQLEGFSAE SRKLLLIVQA LRDNLEPGTF DLEGLYEAIE ECLINDPWVL LNASWFNSFL THALR UniProtKB: Polymerase acidic protein |
-Macromolecule #2: RNA-directed RNA polymerase catalytic subunit
| Macromolecule | Name: RNA-directed RNA polymerase catalytic subunit / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Zhejiang/DTID-ZJU01/2013(H7N9)) Influenza A virus (A/Zhejiang/DTID-ZJU01/2013(H7N9))Strain: A/Zhejiang/DTID-ZJU01/2013(H7N9) |
| Molecular weight | Theoretical: 86.496156 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDVNPTLLFL KVPVQNAIST TFPYTGDPPY SHGTGTGYTM DTVNRTHKYS EKGKWTTNTE TGAPQLNPID GPLPEDNEPS GYAQTDCVL EAMAFLEESH PGIFENSCLE TMEIVQQTRV DKLTQGRQTY DWTLNRNQPA ATALANTIEV FRSNGLTANE S GRLIDFLK ...String: MDVNPTLLFL KVPVQNAIST TFPYTGDPPY SHGTGTGYTM DTVNRTHKYS EKGKWTTNTE TGAPQLNPID GPLPEDNEPS GYAQTDCVL EAMAFLEESH PGIFENSCLE TMEIVQQTRV DKLTQGRQTY DWTLNRNQPA ATALANTIEV FRSNGLTANE S GRLIDFLK DVMDSMDKEE MEITTHFQRK RRVRDNMTKK MVTQRTIGKK KQRLNKRSYL IRALTLNTMT KDAERGKLKR RA IATPGMQ IRGFVYFVEA LARSICEKLE QSGLPVGGNE KKAKLANVVR KMMTNSQDTE LSFTITGDNT KWNENQNPRM FLA MITYIT RNQPEWFRNV LSIAPIMFSN KMARLGKGYM FESKSMKLRT QVPAEMLANI DLKYFNKSTR EKIEKIRPLL IDGT ASLSP GMMMGMFNML STVLGVSILN LGQKKYTKTT YWWDGLQSSD DFALIVNAPN HEGIQAGVDR FYRTCKLVGI NMSKK KSYI NRTGTFEFTS FFYRYGFVAN FSMELPSFGV SGINESADMS VGVTVIKNNM INNDLGPATA QMALQLFIKD YRYTYR CHR GDTQIQTRRA FELKKLWEQT RSKAGLLVSD GGPNLYNIRN LHIPEVCLKW ELMDEDYQGR LCNPMNPFVS HKEIDSV NN AVVMPAHGPA KSMEYDAVAT THSWIPKRNR SILNTSQRGI LEDEQMYQKC CNLFEKFFPS SSYRRPVGIS SMVEAMVS R ARIDARIDFE SGRIKKEEFA EIMKICSTIE ELRRQK UniProtKB: RNA-directed RNA polymerase catalytic subunit |
-Macromolecule #3: Polymerase basic protein 2
| Macromolecule | Name: Polymerase basic protein 2 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Zhejiang/DTID-ZJU01/2013(H7N9)) Influenza A virus (A/Zhejiang/DTID-ZJU01/2013(H7N9))Strain: A/Zhejiang/DTID-ZJU01/2013(H7N9) |
| Molecular weight | Theoretical: 86.00732 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MERIKELRDL MSQSRTREIL TKTTVDHMAI IKKYTSGRQE KNPALRMKWM MAMKYPITAD KRIMEMIPER NEQGQTLWSK TNDAGSDRV MVSPLAVTWW NRNGPTTSTV HYPKVYKTYF EKVERLKHGT FGPVHFRNQV KIRRRVDINP GHADLSAKEA Q DVIMEVVF ...String: MERIKELRDL MSQSRTREIL TKTTVDHMAI IKKYTSGRQE KNPALRMKWM MAMKYPITAD KRIMEMIPER NEQGQTLWSK TNDAGSDRV MVSPLAVTWW NRNGPTTSTV HYPKVYKTYF EKVERLKHGT FGPVHFRNQV KIRRRVDINP GHADLSAKEA Q DVIMEVVF PNEVGARILT SESQLTITKE KKKELQDCKI APLMVAYMLE RELVRKTRFL PVAGGTSSVY IEVLHLTQGT CW EQMYTPG GEVRNDDVDQ SLIIAARNIV RRATVSADPL ASLLEMCHST QIGGVRMVDI LRQNPTEEQA VDICKAAMGL RIS SSFSFG GFTFKRTSGS SVKREEEVLT GNLQTLKIRV HEGYEEFTMV GRRATAILRK ATRRLIQLIV SGKDEQSIAE AIIV AMVFS QEDCMIKAVR GDLNFVNRAN QRLNPMHQLL RHFQKDAKVL FQNWGIEPID NVMGMIGILP DMTPSTEMSL RGVRV SKMG VDEYSSTERV VVSIDRFLRV RDQRGNVLLS PEEVSETQGT EKLTITYSSS MMWEINGPES VLVNTYQWII RNWENV KIQ WSQDPTMLYN KMEFEPFQSL VPKAARGQYS GFVRVLFQQM RDVLGTFDTV QIIKLLPFAA APPEQSRMQF SSLTVNV RG SGMRIVVRGN SPVFNYNKAT KRLTVLGKDA GALMEDPDEG TAGVESAVLR GFLILGKENK RYGPALSINE LSNLAKGE K ANVLIGQGDV VLVMKRKRDS SILTDSQTAT KRIRMAIN UniProtKB: Polymerase basic protein 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)