[English] 日本語
 Yorodumi
Yorodumi- EMDB-14847: Cryo-EM structure of a CRISPR effector in complex with regulator -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a CRISPR effector in complex with regulator | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-cas / effector / regulator / CrRNA / guide RNA / antiphage / ANTIVIRAL PROTEIN | |||||||||
| Biological species |  Desulfonema magnum (bacteria) Desulfonema magnum (bacteria) | |||||||||
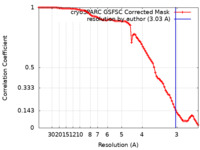
| Method | single particle reconstruction / cryo EM / Resolution: 3.03 Å | |||||||||
 Authors Authors | Babatunde EE / Davide T / Bertrand B / Sergey N / Alexander M / Henning S / Dong CN | |||||||||
| Funding support |  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structural insights into the regulation of Cas7-11 by TPR-CHAT. Authors: Babatunde Ekundayo / Davide Torre / Bertrand Beckert / Sergey Nazarov / Alexander Myasnikov / Henning Stahlberg / Dongchun Ni /  Abstract: The CRISPR-guided caspase (Craspase) complex is an assembly of the target-specific RNA nuclease known as Cas7-11 bound to CRISPR RNA (crRNA) and an ancillary protein known as TPR-CHAT ...The CRISPR-guided caspase (Craspase) complex is an assembly of the target-specific RNA nuclease known as Cas7-11 bound to CRISPR RNA (crRNA) and an ancillary protein known as TPR-CHAT (tetratricopeptide repeats (TPR) fused with a CHAT domain). The Craspase complex holds promise as a tool for gene therapy and biomedical research, but its regulation is poorly understood. TPR-CHAT regulates Cas7-11 nuclease activity via an unknown mechanism. In the present study, we use cryoelectron microscopy to determine structures of the Desulfonema magnum (Dm) Craspase complex to gain mechanistic insights into its regulation. We show that DmTPR-CHAT stabilizes crRNA-bound DmCas7-11 in a closed conformation via a network of interactions mediated by the DmTPR-CHAT N-terminal domain, the DmCas7-11 insertion finger and Cas11-like domain, resulting in reduced target RNA accessibility and cleavage. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14847.map.gz emd_14847.map.gz | 63.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14847-v30.xml emd-14847-v30.xml emd-14847.xml emd-14847.xml | 25.8 KB 25.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14847_fsc.xml emd_14847_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_14847.png emd_14847.png | 63.6 KB | ||
| Filedesc metadata |  emd-14847.cif.gz emd-14847.cif.gz | 7.2 KB | ||
| Others |  emd_14847_additional_1.map.gz emd_14847_additional_1.map.gz emd_14847_additional_2.map.gz emd_14847_additional_2.map.gz emd_14847_additional_3.map.gz emd_14847_additional_3.map.gz emd_14847_half_map_1.map.gz emd_14847_half_map_1.map.gz emd_14847_half_map_2.map.gz emd_14847_half_map_2.map.gz | 34 MB 63.4 MB 63.3 MB 62.3 MB 62.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14847 http://ftp.pdbj.org/pub/emdb/structures/EMD-14847 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14847 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14847 | HTTPS FTP |
-Validation report
| Summary document |  emd_14847_validation.pdf.gz emd_14847_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14847_full_validation.pdf.gz emd_14847_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_14847_validation.xml.gz emd_14847_validation.xml.gz | 16.4 KB | Display | |
| Data in CIF |  emd_14847_validation.cif.gz emd_14847_validation.cif.gz | 20.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14847 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14847 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14847 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14847 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14847.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14847.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2769 Å | ||||||||||||||||||||||||||||||||||||
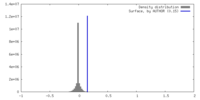
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #3
| File | emd_14847_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
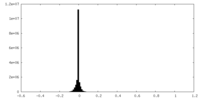
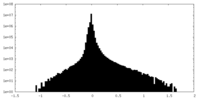
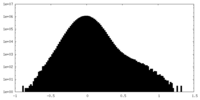
| Density Histograms |
-Additional map: #2
| File | emd_14847_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
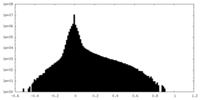
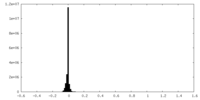
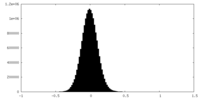
| Density Histograms |
-Additional map: #1
| File | emd_14847_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
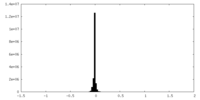
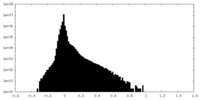
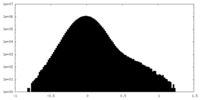
| Density Histograms |
-Half map: #2
| File | emd_14847_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
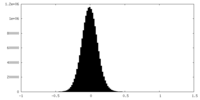
| Density Histograms |
-Half map: #1
| File | emd_14847_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cas7-11 with TRP-CHAT-NTD
| Entire | Name: Cas7-11 with TRP-CHAT-NTD |
|---|---|
| Components |
|
-Supramolecule #1: Cas7-11 with TRP-CHAT-NTD
| Supramolecule | Name: Cas7-11 with TRP-CHAT-NTD / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Desulfonema magnum (bacteria) Desulfonema magnum (bacteria) |
| Molecular weight | Theoretical: 220 KDa |
-Macromolecule #1: Cas7-11
| Macromolecule | Name: Cas7-11 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Desulfonema magnum (bacteria) Desulfonema magnum (bacteria) |
| Molecular weight | Theoretical: 207.402859 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SQDPMQHIIP LTITFLESFR VIEWHKSSDR NSLRFLRGYA YARWHRSLKN NKGRPYITGT LVRSAIIRAA EELLWLNDG NYKGAICCPG EFNGSNAKVF REGKARRLRR RQTLTWPTKC ACHEKEPCPF CLLLGRYEKS SKTSKSPVLN N VNFSNFNV ...String: MGSSHHHHHH SQDPMQHIIP LTITFLESFR VIEWHKSSDR NSLRFLRGYA YARWHRSLKN NKGRPYITGT LVRSAIIRAA EELLWLNDG NYKGAICCPG EFNGSNAKVF REGKARRLRR RQTLTWPTKC ACHEKEPCPF CLLLGRYEKS SKTSKSPVLN N VNFSNFNV LGKEKEFLNI EDVADMRVVN RVDQQSGKAE DFFNIWEITD GAWKIFRGEI QVSDKGWSDE ENFSKFLTLL KG ASVLVDK ISGGLCYLTL DKPELAELPA VKTEKDVLEP GDDASVLEIS RPPYWNNMLN LAGTISEAFE REDKLVHLRL FAD TVRELR RSDIETLDLP KGHADRLGKP SDHFIWDIEI NKKVKLRNWL FWIFNEFRDT YAYFDWRTFC EALGQALYLE AKKQ VPNQF SSERPVGATP AMEVKTPEHD PGRAAQGPRY EWLIKGELVS QTPFFFGWST EADNREHTNL KLLAARDGRL RLPLS VLRG ILRRDIKVVL DNKCRAELAM KQPCSCPVCN LMKKITIRDS FSSNYAEPPK IRHKIRLDPK SGTVAKGALF DSEVGP RGV VFPFELRLRS ADDTLPQALK TVFSWWQQGT VSFSGDAGTG KGIFCLRNLK SIRWDLKTEM DKYAATLGGR KTPVGKW DK CHIPDDKTYP WVKETVEISV CSPFITKDPV NSLIDSAGYD AICYTTVDLE KSENINSLPE SEISLLFEMF DLQYPMSY L FPNKDIYLLK GESFRGMLRT AVGRGENLLL REHEDCSCTL CRIFGNEHNA GKIRVEDFII QGEPRTKLVD RVAIDRFTA GAKDKFKFDA APIVGTPTNK LKFRGNIWIH RDLDGLACES LKLALEDIEN GLYPFGGLGN AGFGWVNYNP LSHPAQEENK ADFSLTKKM ELNWSMKELS TDKIYWPHYF LPFGKKVLRE KTPPSHACID ENEDSELYSG KIVCTLETRT PLIIPDSEFQ G EEHKSYDF FNLNGELCIP GSEIRGMISS VFEALTNSCM RIFDEKKRLS WRMNPNKKDK KNNNRRELDD FIPGRVTNDR KM EEMKEYR YPFYDQAITA NDKQNKYFDQ WEATIELTDE SLEKLKAEKI LQSVLDALRP LTKKKEKYKN TETFVFDLKK FIG KIDDNQ QMISEALERG KVRLTGNSLK QISKTKKIPR QILNQLKGLK DNTYENREQF ISVLKTTVRG INDEQISLIL DNID EDVRL TDLSLTKIRK AKVPQTLVDM LADLKKDEPY KNEKEFLSEF KKKMGEIIGL ILKHAAKTGG DVPRYNHPTP TDKML LSLA AYNRNHKHEN GKAEYRIVKP KHNLKVDFMF AVTPFENPFK GYNPAAVVEE PVGGYLKVSG PNKIEKVKKV NPNSVS VRD DKNQEIIHNG VYLRKITVAN AKSKNKLRER LVPEFAWYDK DSEAAYAMTK RCERVFVEIG AKPIPIQPSA REKFKIL TQ EYQKNAKQQK TPEAFQTILP KDGELRPGDL VYFREDKKTN TVTDIIPVRI SRTVDDEVLA RKIPDDVGDV RPCVREIL D KEKQKEIADA GVKEVFQHHP DGLCPACSLF GTTFYKGRIA FGFAFHKDKD PELANNGKHI TLPLLERPRP TWSMPKKES RVPGRKFYVH HQGWERVIKH SNLDESDPNA TKQTVNNRSV QAIKEEQKFQ FEVRFENLRE WELGLLVYVL QLEPQFAHKL GMGKALGFG SVRIRVGEIH SGPKELDESR LVSSAMKKME EIWDRDKNVL EKLFRLLYFN ESKDIKVRYP KLQKEKEEEE E ESGYMELA KEEYQPEQRR NKLTNPWEGW GNILKKPAIL I |
-Macromolecule #3: TPR-CHAT
| Macromolecule | Name: TPR-CHAT / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Desulfonema magnum (bacteria) Desulfonema magnum (bacteria) |
| Molecular weight | Theoretical: 17.361045 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSAFSGLKI PELSVDPAEV FKSDNPQLVS VLLDEFELQE QRPFFSGLIP EKQINIALKK SPQLKKLACH LLEAYEINGR RWKHADRRR VLEKAIRLLE KVSNELKGDI QKLENNVKES GKDSEELNKT REKHGEILAD MGRAYLHRAK II |
-Macromolecule #2: gRNA
| Macromolecule | Name: gRNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Desulfonema magnum (bacteria) Desulfonema magnum (bacteria) |
| Molecular weight | Theoretical: 12.344251 KDa |
| Sequence | String: UAUGUGAUGG AACCUCUCCG GAUAAUUCUA UCUCUUCUG |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 43 |
|---|---|
| Output model |  PDB-7zol: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)