[English] 日本語
 Yorodumi
Yorodumi- EMDB-14777: Cryo-EM structure of archaic chaperone-usher Csu pilus of Acineto... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of archaic chaperone-usher Csu pilus of Acinetobacter baumannii | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology | Spore coat protein U / Spore Coat Protein U domain / Spore Coat Protein U domain / Uncharacterized protein Function and homology information Function and homology information | |||||||||
| Biological species |   Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) | |||||||||
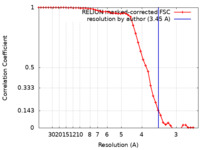
| Method | helical reconstruction /  cryo EM / Resolution: 3.45 Å cryo EM / Resolution: 3.45 Å | |||||||||
 Authors Authors | Pakharukova N / Malmi H / Tuittila M / Paavilainen S / Ghosal D / Chang YW / Jensen GJ / Zavialov AV | |||||||||
| Funding support |  Finland, 2 items Finland, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Archaic chaperone-usher pili self-secrete into superelastic zigzag springs. Authors: Natalia Pakharukova / Henri Malmi / Minna Tuittila / Tobias Dahlberg / Debnath Ghosal / Yi-Wei Chang / Si Lhyam Myint / Sari Paavilainen / Stefan David Knight / Urpo Lamminmäki / Bernt Eric ...Authors: Natalia Pakharukova / Henri Malmi / Minna Tuittila / Tobias Dahlberg / Debnath Ghosal / Yi-Wei Chang / Si Lhyam Myint / Sari Paavilainen / Stefan David Knight / Urpo Lamminmäki / Bernt Eric Uhlin / Magnus Andersson / Grant Jensen / Anton V Zavialov /     Abstract: Adhesive pili assembled through the chaperone-usher pathway are hair-like appendages that mediate host tissue colonization and biofilm formation of Gram-negative bacteria. Archaic chaperone-usher ...Adhesive pili assembled through the chaperone-usher pathway are hair-like appendages that mediate host tissue colonization and biofilm formation of Gram-negative bacteria. Archaic chaperone-usher pathway pili, the most diverse and widespread chaperone-usher pathway adhesins, are promising vaccine and drug targets owing to their prevalence in the most troublesome multidrug-resistant pathogens. However, their architecture and assembly-secretion process remain unknown. Here, we present the cryo-electron microscopy structure of the prototypical archaic Csu pilus that mediates biofilm formation of Acinetobacter baumannii-a notorious multidrug-resistant nosocomial pathogen. In contrast to the thick helical tubes of the classical type 1 and P pili, archaic pili assemble into an ultrathin zigzag architecture secured by an elegant clinch mechanism. The molecular clinch provides the pilus with high mechanical stability as well as superelasticity, a property observed for the first time, to our knowledge, in biomolecules, while enabling a more economical and faster pilus production. Furthermore, we demonstrate that clinch formation at the cell surface drives pilus secretion through the outer membrane. These findings suggest that clinch-formation inhibitors might represent a new strategy to fight multidrug-resistant bacterial infections. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14777.map.gz emd_14777.map.gz | 5.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14777-v30.xml emd-14777-v30.xml emd-14777.xml emd-14777.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14777_fsc.xml emd_14777_fsc.xml | 4.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_14777.png emd_14777.png | 73.5 KB | ||
| Others |  emd_14777_half_map_1.map.gz emd_14777_half_map_1.map.gz emd_14777_half_map_2.map.gz emd_14777_half_map_2.map.gz | 6 MB 6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14777 http://ftp.pdbj.org/pub/emdb/structures/EMD-14777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14777 | HTTPS FTP |
-Related structure data
| Related structure data |  7zl4MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14777.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14777.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2949 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half-map 2
| File | emd_14777_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1
| File | emd_14777_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Csu pilus of Acinetobacter baumannii
| Entire | Name: Csu pilus of Acinetobacter baumannii |
|---|---|
| Components |
|
-Supramolecule #1: Csu pilus of Acinetobacter baumannii
| Supramolecule | Name: Csu pilus of Acinetobacter baumannii / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #1: CsuA/B
| Macromolecule | Name: CsuA/B / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Molecular weight | Theoretical: 16.069642 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: AVTGQVDVKL NISTGCTVGG SQTEGNMNKF GTLNFGKTSG TWNNVLTAEV ASAATGGNIS VTCDGTDPVD FTVAIDGGER TDRTLKNTA SADVVAYNVY RDAARTNLYV VNQPQQFTTV SGQATAVPIF GAIAPNTGTP KAQGDYKDTL LVTVNF |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)