+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse heavy chain apoferritin in plunge-frozen vitrified ice | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.57 Å | |||||||||
 Authors Authors | Harder OF / Voss JM / Olshin PK / Drabbels M / Lorenz UJ | |||||||||
| Funding support | European Union,  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: Acta Crystallogr D Struct Biol / Year: 2022 Journal: Acta Crystallogr D Struct Biol / Year: 2022Title: Microsecond melting and revitrification of cryo samples: protein structure and beam-induced motion. Authors: Oliver F Harder / Jonathan M Voss / Pavel K Olshin / Marcel Drabbels / Ulrich J Lorenz /  Abstract: A novel approach to time-resolved cryo-electron microscopy (cryo-EM) has recently been introduced that involves melting a cryo sample with a laser beam to allow protein dynamics to briefly occur in ...A novel approach to time-resolved cryo-electron microscopy (cryo-EM) has recently been introduced that involves melting a cryo sample with a laser beam to allow protein dynamics to briefly occur in the liquid, before trapping the particles in their transient configurations by rapidly revitrifying the sample. With a time resolution of just a few microseconds, this approach is notably fast enough to study the domain motions that are typically associated with the activity of proteins but which have previously remained inaccessible. Here, crucial details are added to the characterization of the method. It is shown that single-particle reconstructions of apoferritin and Cowpea chlorotic mottle virus from revitrified samples are indistinguishable from those from conventional samples, demonstrating that melting and revitrification leaves the particles intact and that they do not undergo structural changes within the spatial resolution afforded by the instrument. How rapid revitrification affects the properties of the ice is also characterized, showing that revitrified samples exhibit comparable amounts of beam-induced motion. The results pave the way for microsecond time-resolved studies of the conformational dynamics of proteins and open up new avenues to study the vitrification process and to address beam-induced specimen movement. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14767.map.gz emd_14767.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14767-v30.xml emd-14767-v30.xml emd-14767.xml emd-14767.xml | 11.8 KB 11.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14767.png emd_14767.png | 112.6 KB | ||
| Others |  emd_14767_half_map_1.map.gz emd_14767_half_map_1.map.gz emd_14767_half_map_2.map.gz emd_14767_half_map_2.map.gz | 28.1 MB 28.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14767 http://ftp.pdbj.org/pub/emdb/structures/EMD-14767 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14767 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14767 | HTTPS FTP |
-Validation report
| Summary document |  emd_14767_validation.pdf.gz emd_14767_validation.pdf.gz | 805.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14767_full_validation.pdf.gz emd_14767_full_validation.pdf.gz | 805.5 KB | Display | |
| Data in XML |  emd_14767_validation.xml.gz emd_14767_validation.xml.gz | 10.5 KB | Display | |
| Data in CIF |  emd_14767_validation.cif.gz emd_14767_validation.cif.gz | 12.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14767 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14767 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14767 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14767 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| EM raw data |  EMPIAR-11016 (Title: Single particle cryo-EM dataset of mouse heavy chain apoferritin in plunge-frozen vitrified ice EMPIAR-11016 (Title: Single particle cryo-EM dataset of mouse heavy chain apoferritin in plunge-frozen vitrified iceData size: 82.8 Data #1: Unaligned multi-frame micrographs of mouse heavy chain apoferritin in plunge-frozen vitrified ice [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14767.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14767.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.975 Å | ||||||||||||||||||||||||||||||||||||
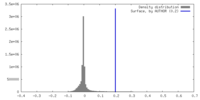
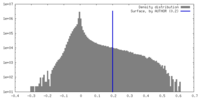
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_14767_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
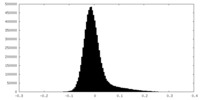
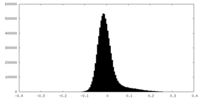
| Density Histograms |
-Half map: #2
| File | emd_14767_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
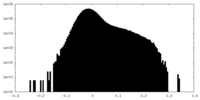
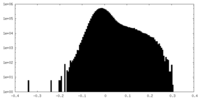
| Density Histograms |
- Sample components
Sample components
-Entire : Mouse heavy chain apoferritin in plunge-frozen vitrified ice
| Entire | Name: Mouse heavy chain apoferritin in plunge-frozen vitrified ice |
|---|---|
| Components |
|
-Supramolecule #1: Mouse heavy chain apoferritin in plunge-frozen vitrified ice
| Supramolecule | Name: Mouse heavy chain apoferritin in plunge-frozen vitrified ice type: complex / Chimera: Yes / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.57 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 49885 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)