+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Amyloid fibril (in vitro) from full-length hnRNPA1 protein | |||||||||
 Map data Map data | Final map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Amyloidosis / Misfolding disease / Amyloidosis / Misfolding disease /  Inflammation / Inflammation /  Prion / Protein fibril / Prion / Protein fibril /  NUCLEAR PROTEIN NUCLEAR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to sodium arsenite / SARS-CoV-1-host interactions / import into nucleus /  telomeric repeat-containing RNA binding / G-rich strand telomeric DNA binding / telomeric repeat-containing RNA binding / G-rich strand telomeric DNA binding /  pre-mRNA binding / pre-mRNA binding /  nuclear export / RNA export from nucleus / miRNA binding / FGFR2 alternative splicing ...cellular response to sodium arsenite / SARS-CoV-1-host interactions / import into nucleus / nuclear export / RNA export from nucleus / miRNA binding / FGFR2 alternative splicing ...cellular response to sodium arsenite / SARS-CoV-1-host interactions / import into nucleus /  telomeric repeat-containing RNA binding / G-rich strand telomeric DNA binding / telomeric repeat-containing RNA binding / G-rich strand telomeric DNA binding /  pre-mRNA binding / pre-mRNA binding /  nuclear export / RNA export from nucleus / miRNA binding / FGFR2 alternative splicing / regulation of alternative mRNA splicing, via spliceosome / intracellular non-membrane-bounded organelle / SARS-CoV-1 modulates host translation machinery / nuclear export / RNA export from nucleus / miRNA binding / FGFR2 alternative splicing / regulation of alternative mRNA splicing, via spliceosome / intracellular non-membrane-bounded organelle / SARS-CoV-1 modulates host translation machinery /  regulation of RNA splicing / negative regulation of telomere maintenance via telomerase / Processing of Capped Intron-Containing Pre-mRNA / mRNA transport / localization / cellular response to glucose starvation / positive regulation of telomere maintenance via telomerase / catalytic step 2 spliceosome / molecular condensate scaffold activity / mRNA Splicing - Major Pathway / mRNA 3'-UTR binding / regulation of RNA splicing / negative regulation of telomere maintenance via telomerase / Processing of Capped Intron-Containing Pre-mRNA / mRNA transport / localization / cellular response to glucose starvation / positive regulation of telomere maintenance via telomerase / catalytic step 2 spliceosome / molecular condensate scaffold activity / mRNA Splicing - Major Pathway / mRNA 3'-UTR binding /  spliceosomal complex / spliceosomal complex /  mRNA splicing, via spliceosome / mRNA splicing, via spliceosome /  single-stranded DNA binding / amyloid fibril formation / single-stranded DNA binding / amyloid fibril formation /  single-stranded RNA binding / single-stranded RNA binding /  ribonucleoprotein complex / protein domain specific binding / ribonucleoprotein complex / protein domain specific binding /  DNA binding / DNA binding /  RNA binding / extracellular exosome / RNA binding / extracellular exosome /  nucleoplasm / nucleoplasm /  membrane / identical protein binding / membrane / identical protein binding /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
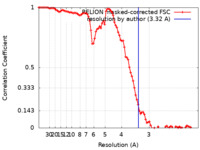
| Method | helical reconstruction /  cryo EM / Resolution: 3.32 Å cryo EM / Resolution: 3.32 Å | |||||||||
 Authors Authors | Sharma K / Banerjee S / Schmidt M / Faendrich M | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2023 Journal: J Mol Biol / Year: 2023Title: Cryo-EM Structure of the Full-length hnRNPA1 Amyloid Fibril. Authors: Kartikay Sharma / Sambhasan Banerjee / Dilan Savran / Cedric Rajes / Sebastian Wiese / Amandeep Girdhar / Nadine Schwierz / Christopher Lee / James Shorter / Matthias Schmidt / Lin Guo / Marcus Fändrich /   Abstract: Heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) is a multifunctional RNA-binding protein that is associated with neurodegenerative diseases, such as amyotrophic lateral sclerosis and multisystem ...Heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) is a multifunctional RNA-binding protein that is associated with neurodegenerative diseases, such as amyotrophic lateral sclerosis and multisystem proteinopathy. In this study, we have used cryo-electron microscopy to investigate the three-dimensional structure of amyloid fibrils from full-length hnRNPA1 protein. We find that the fibril core is formed by a 45-residue segment of the prion-like low-complexity domain of the protein, whereas the remaining parts of the protein (275 residues) form a fuzzy coat around the fibril core. The fibril consists of two fibril protein stacks that are arranged into a pseudo-2 screw symmetry. The ordered core harbors several of the positions that are known to be affected by disease-associated mutations, but does not encompass the most aggregation-prone segments of the protein. These data indicate that the structures of amyloid fibrils from full-length proteins may be more complex than anticipated by current theories on protein misfolding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14739.map.gz emd_14739.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14739-v30.xml emd-14739-v30.xml emd-14739.xml emd-14739.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14739_fsc.xml emd_14739_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_14739.png emd_14739.png | 71.2 KB | ||
| Others |  emd_14739_half_map_1.map.gz emd_14739_half_map_1.map.gz emd_14739_half_map_2.map.gz emd_14739_half_map_2.map.gz | 49.2 MB 49.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14739 http://ftp.pdbj.org/pub/emdb/structures/EMD-14739 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14739 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14739 | HTTPS FTP |
-Related structure data
| Related structure data |  7zj2MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14739.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14739.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
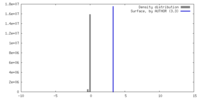
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Relion half map-1
| File | emd_14739_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion half map-1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
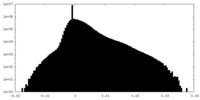
| Density Histograms |
-Half map: Relion half map-2
| File | emd_14739_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion half map-2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Amyloid fibril (in vitro) from full-length hnRNPA1 protein
| Entire | Name: Amyloid fibril (in vitro) from full-length hnRNPA1 protein |
|---|---|
| Components |
|
-Supramolecule #1: Amyloid fibril (in vitro) from full-length hnRNPA1 protein
| Supramolecule | Name: Amyloid fibril (in vitro) from full-length hnRNPA1 protein type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: In vitro full-length hnRNPA1 amyloid fibril |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Isoform A1-A of Heterogeneous nuclear ribonucleoprotein A1
| Macromolecule | Name: Isoform A1-A of Heterogeneous nuclear ribonucleoprotein A1 type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.246227 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MSKSESPKEP EQLRKLFIGG LSFETTDESL RSHFEQWGTL TDCVVMRDPN TKRSRGFGFV TYATVEEVDA AMNARPHKVD GRVVEPKRA VSREDSQRPG AHLTVKKIFV GGIKEDTEEH HLRDYFEQYG KIEVIEIMTD RGSGKKRGFA FVTFDDHDSV D KIVIQKYH ...String: MSKSESPKEP EQLRKLFIGG LSFETTDESL RSHFEQWGTL TDCVVMRDPN TKRSRGFGFV TYATVEEVDA AMNARPHKVD GRVVEPKRA VSREDSQRPG AHLTVKKIFV GGIKEDTEEH HLRDYFEQYG KIEVIEIMTD RGSGKKRGFA FVTFDDHDSV D KIVIQKYH TVNGHNCEVR KALSKQEMAS ASSSQRGRSG SGNFGGGRGG GFGGNDNFGR GGNFSGRGGF GGSRGGGGYG GS GDGYNGF GNDGSNFGGG GSYNDFGNYN NQSSNFGPMK GGNFGGRSSG PYGGGGQYFA KPRNQGGYGG SSSSSSYGSG RRF UniProtKB: Heterogeneous nuclear ribonucleoprotein A1 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 2624 / Average exposure time: 10.0 sec. / Average electron dose: 42.64 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)