[English] 日本語
 Yorodumi
Yorodumi- EMDB-14710: Polymerase module of CPF in complex with Mpe1 and a pre-cleaved C... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Polymerase module of CPF in complex with Mpe1 and a pre-cleaved CYC1 RNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CPF / 3'-end processing / polyA / RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information: / Processing of Intronless Pre-mRNAs / termination of RNA polymerase II transcription, poly(A)-coupled / mRNA cleavage and polyadenylation specificity factor complex / mRNA 3'-end processing / termination of RNA polymerase II transcription / mRNA processing / ubiquitin protein ligase activity / ubiquitin-dependent protein catabolic process / nucleic acid binding ...: / Processing of Intronless Pre-mRNAs / termination of RNA polymerase II transcription, poly(A)-coupled / mRNA cleavage and polyadenylation specificity factor complex / mRNA 3'-end processing / termination of RNA polymerase II transcription / mRNA processing / ubiquitin protein ligase activity / ubiquitin-dependent protein catabolic process / nucleic acid binding / protein ubiquitination / mitochondrion / RNA binding / zinc ion binding / nucleus / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Rodriguez-Molina JB / Passmore LA | |||||||||
| Funding support | European Union,  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Mpe1 senses the binding of pre-mRNA and controls 3' end processing by CPF. Authors: Juan B Rodríguez-Molina / Francis J O'Reilly / Holly Fagarasan / Eleanor Sheekey / Sarah Maslen / J Mark Skehel / Juri Rappsilber / Lori A Passmore /   Abstract: Most eukaryotic messenger RNAs (mRNAs) are processed at their 3' end by the cleavage and polyadenylation specificity factor (CPF/CPSF). CPF mediates the endonucleolytic cleavage of the pre-mRNA and ...Most eukaryotic messenger RNAs (mRNAs) are processed at their 3' end by the cleavage and polyadenylation specificity factor (CPF/CPSF). CPF mediates the endonucleolytic cleavage of the pre-mRNA and addition of a polyadenosine (poly(A)) tail, which together define the 3' end of the mature transcript. The activation of CPF is highly regulated to maintain the fidelity of RNA processing. Here, using cryo-EM of yeast CPF, we show that the Mpe1 subunit directly contacts the polyadenylation signal sequence in nascent pre-mRNA. The region of Mpe1 that contacts RNA also promotes the activation of CPF endonuclease activity and controls polyadenylation. The Cft2 subunit of CPF antagonizes the RNA-stabilized configuration of Mpe1. In vivo, the depletion or mutation of Mpe1 leads to widespread defects in transcription termination by RNA polymerase II, resulting in transcription interference on neighboring genes. Together, our data suggest that Mpe1 plays a major role in accurate 3' end processing, activating CPF, and ensuring timely transcription termination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14710.map.gz emd_14710.map.gz | 150.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14710-v30.xml emd-14710-v30.xml emd-14710.xml emd-14710.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14710.png emd_14710.png | 92 KB | ||
| Filedesc metadata |  emd-14710.cif.gz emd-14710.cif.gz | 7.9 KB | ||
| Others |  emd_14710_half_map_1.map.gz emd_14710_half_map_1.map.gz emd_14710_half_map_2.map.gz emd_14710_half_map_2.map.gz | 127.2 MB 127.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14710 http://ftp.pdbj.org/pub/emdb/structures/EMD-14710 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14710 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14710 | HTTPS FTP |
-Validation report
| Summary document |  emd_14710_validation.pdf.gz emd_14710_validation.pdf.gz | 989.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14710_full_validation.pdf.gz emd_14710_full_validation.pdf.gz | 989 KB | Display | |
| Data in XML |  emd_14710_validation.xml.gz emd_14710_validation.xml.gz | 14.5 KB | Display | |
| Data in CIF |  emd_14710_validation.cif.gz emd_14710_validation.cif.gz | 17.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14710 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14710 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14710 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14710 | HTTPS FTP |
-Related structure data
| Related structure data |  7zgpMC  7zgqC  7zgrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14710.map.gz / Format: CCP4 / Size: 160.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14710.map.gz / Format: CCP4 / Size: 160.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
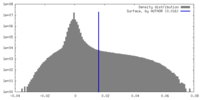
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_14710_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
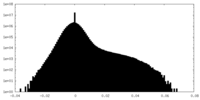
| Density Histograms |
-Half map: #1
| File | emd_14710_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : polymerase module-Mpe1-RNA
| Entire | Name: polymerase module-Mpe1-RNA |
|---|---|
| Components |
|
-Supramolecule #1: polymerase module-Mpe1-RNA
| Supramolecule | Name: polymerase module-Mpe1-RNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Protein CFT1
| Macromolecule | Name: Protein CFT1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 153.577156 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNVYDDVLDA TVVSHSLATH FTTSDYEELL VVRTNILSVY RPTRDGKLYL TDEFKFHGLI TDIGLIPQKD SPLSCLLLCT GVAKISILK FNTLTNSIDT LSLHYYEGKF KGKSLVELAK ISTLRMDPGS SCALLFNNDI IAFLPFHVNK NDDDEEEEDE D ENIDDSEL ...String: MNVYDDVLDA TVVSHSLATH FTTSDYEELL VVRTNILSVY RPTRDGKLYL TDEFKFHGLI TDIGLIPQKD SPLSCLLLCT GVAKISILK FNTLTNSIDT LSLHYYEGKF KGKSLVELAK ISTLRMDPGS SCALLFNNDI IAFLPFHVNK NDDDEEEEDE D ENIDDSEL IHSMNQKSQG TNTFNKRKRT KLGDKFTAPS VVLVASELYE GAKNIIDIQF LKNFTKPTIA LLYQPKLVWA GN TTISKLP TQYVILTLNI QPAESATKIE STTIAFVKEL PWDLHTIVPV SNGAIIVGTN ELAFLDNTGV LQSTVLLNSF ADK ELQKTK IINNSSLEIM FREKNTTSIW IPSSKSKNGG SNNDETLLLM DLKSNIYYIQ MEAEGRLLIK FDIFKLPIVN DLLK ENSNP KCITRLNATN SNKNMDLFIG FGSGNALVLR LNNLKSTIET REAHNPSSGT NSLMDINDDD DEEMDDLYAD EAPEN GLTT NDSKGTVETV QPFDIELLSS LRNVGPITSL TVGKVSSIDD VVKGLPNPNK NEYSLVATSG NGSGSHLTVI QTSVQP EIE LALKFISITQ IWNLKIKGRD RYLITTDSTK SRSDIYESDN NFKLHKGGRL RRDATTVYIS MFGEEKRIIQ VTTNHLY LY DTHFRRLTTI KFDYEVIHVS VMDPYILVTV SRGDIKIFEL EEKNKRKLLK VDLPEILNEM VITSGLILKS NMCNEFLI G LSKSQEEQLL FTFVTADNQI IFFTKDHNDR IFQLNGVDQL NESLYISTYQ LGDEIVPDPS IKQVMINKLG HDNKEEYLT ILTFGGEIYQ YRKLPQRRSR FYRNVTRNDL AITGAPDNAY AKGVSSIERI MHYFPDYNGY SVIFVTGSVP YILIKEDDST PKIFKFGNI PLVSVTPWSE RSVMCVDDIK NARVYTLTTD NMYYGNKLPL KQIKISNVLD DYKTLQKLVY HERAQLFLVS Y CKRVPYEA LGEDGEKVIG YDENVPHAEG FQSGILLINP KSWKVIDKID FPKNSVVNEM RSSMIQINSK TKRKREYIIA GV ANATTED TPPTGAFHIY DVIEVVPEPG KPDTNYKLKE IFQEEVSGTV STVCEVSGRF MISQSQKVLV RDIQEDNSVI PVA FLDIPV FVTDSKSFGN LLIIGDAMQG FQFIGFDAEP YRMISLGRSM SKFQTMSLEF LVNGGDMYFA ATDADRNVHV LKYA PDEPN SLSGQRLVHC SSFTLHSTNS CMMLLPRNEE FGSPQVPSFQ NVGGQVDGSV FKIVPLSEEK YRRLYVIQQQ IIDRE LQLG GLNPRMERLA NDFYQMGHSM RPMLDFNVIR RFCGLAIDRR KSIAQKAGRH AHFEAWRDII NIEFSMRSLC QGK UniProtKB: Protein CFT1 |
-Macromolecule #2: mRNA 3'-end-processing protein YTH1
| Macromolecule | Name: mRNA 3'-end-processing protein YTH1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.594498 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSLIHPDTAK YPFKFEPFLR QEYSFSLDPD RPICEFYNSR EGPKSCPRGP LCPKKHVLPI FQNKIVCRHW LRGLCKKNDQ CEYLHEYNL RKMPECVFFS KNGYCTQSPD CQYLHIDPAS KIPKCENYEM GFCPLGSSCP RRHIKKVFCQ RYMTGFCPLG K DECDMEHP ...String: MSLIHPDTAK YPFKFEPFLR QEYSFSLDPD RPICEFYNSR EGPKSCPRGP LCPKKHVLPI FQNKIVCRHW LRGLCKKNDQ CEYLHEYNL RKMPECVFFS KNGYCTQSPD CQYLHIDPAS KIPKCENYEM GFCPLGSSCP RRHIKKVFCQ RYMTGFCPLG K DECDMEHP QFIIPDEGSK LRIKRDDEIN TRKMDEEKER RLNAIINGEV UniProtKB: mRNA 3'-end-processing protein |
-Macromolecule #3: Polyadenylation factor subunit 2
| Macromolecule | Name: Polyadenylation factor subunit 2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.211117 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDGHNQNQYQ NQNQIQQSQQ PPLKKYVTQR RSVDVSSPYI NLYYNRRHGL PNLVVEPETS YTIDIMPPNA YRGRDRVINL PSKFTHLSS NKVKHVIPAI QWTPEGRRLV VATYSGEFSL WNASSFTFET LMQAHDSAVT TMKYSHDSDW MISGDADGMI K IWQPNFSM ...String: MDGHNQNQYQ NQNQIQQSQQ PPLKKYVTQR RSVDVSSPYI NLYYNRRHGL PNLVVEPETS YTIDIMPPNA YRGRDRVINL PSKFTHLSS NKVKHVIPAI QWTPEGRRLV VATYSGEFSL WNASSFTFET LMQAHDSAVT TMKYSHDSDW MISGDADGMI K IWQPNFSM VKEIDAAHTE SIRDMAFSSN DSKFVTCSDD NILKIWNFSN GKQERVLSGH HWDVKSCDWH PEMGLIASAS KD NLVKLWD PRSGNCISSI LKFKHTVLKT RFQPTKGNLL MAISKDKSCR VFDIRYSMKE LMCVRDETDY MTLEWHPINE SMF TLACYD GSLKHFDLLQ NLNEPILTIP YAHDKCITSL SYNPVGHIFA TAAKDRTIRF WTRARPIDPN AYDDPTYNNK KING WFFGI NNDINAVREK SEFGAAPPPP ATLEPHALPN MNGFINKKPR QEIPGIDSNI KSSTLPGLSI UniProtKB: Polyadenylation factor subunit 2 |
-Macromolecule #5: MPE1 isoform 1
| Macromolecule | Name: MPE1 isoform 1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.711848 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSTIFYRFK SQRNTSRILF DGTGLTVFDL KREIIQENKL GDGTDFQLKI YNPDTEEEYD DDAFVIPRST SVIVKRSPAI KSFSVHSRL KGNVGAAALG NATRYVTGRP RVLQKRQHTA TTTANVSGTT EEERIASMFA TQENQWEQTQ EEMSAATPVF F KSQTNKNS ...String: MSSTIFYRFK SQRNTSRILF DGTGLTVFDL KREIIQENKL GDGTDFQLKI YNPDTEEEYD DDAFVIPRST SVIVKRSPAI KSFSVHSRL KGNVGAAALG NATRYVTGRP RVLQKRQHTA TTTANVSGTT EEERIASMFA TQENQWEQTQ EEMSAATPVF F KSQTNKNS AQENEGPPPP GYMCYRCGGR DHWIKNCPTN SDPNFEGKRI RRTTGIPKKF LKSIEIDPET MTPEEMAQRK IM ITDEGKF VVQVEDKQSW EDYQRKRENR QIDGDETIWR KGHFKDLPDD LKCPLTGGLL RQPVKTSKCC NIDFSKEALE NAL VESDFV CPNCETRDIL LDSLVPDQDK EKEVETFLKK QEELHGSSKD GNQPETKKMK LMDPTGTAGL NNNTSLPTSV NNGG TPVPP VPLPFGIPPF PMFPMPFMPP TATITNPHQA DASPKK UniProtKB: MPE1 isoform 1 |
-Macromolecule #4: pre-cleaved CYC1
| Macromolecule | Name: pre-cleaved CYC1 / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.329796 KDa |
| Sequence | String: UUUAUAGUUA UGUUAGUAUU AAGAACGUUA UUUAUAUUUC AA |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: 20 mM HEPES pH 8, 50 mM NaCl, 0.5 mM TCEP |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 11856 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.1 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


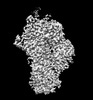



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)