+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | pMMO three trimer interaction map from native membrane | |||||||||
 Map data Map data | pMMO trimer-trimer map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Methylococcus capsulatus str. Bath (bacteria) Methylococcus capsulatus str. Bath (bacteria) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 12.0 Å | |||||||||
 Authors Authors | Zhu Y / Ni T / Zhang P | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure and activity of particulate methane monooxygenase arrays in methanotrophs. Authors: Yanan Zhu / Christopher W Koo / C Keith Cassidy / Matthew C Spink / Tao Ni / Laura C Zanetti-Domingues / Benji Bateman / Marisa L Martin-Fernandez / Juan Shen / Yuewen Sheng / Yun Song / ...Authors: Yanan Zhu / Christopher W Koo / C Keith Cassidy / Matthew C Spink / Tao Ni / Laura C Zanetti-Domingues / Benji Bateman / Marisa L Martin-Fernandez / Juan Shen / Yuewen Sheng / Yun Song / Zhengyi Yang / Amy C Rosenzweig / Peijun Zhang /    Abstract: Methane-oxidizing bacteria play a central role in greenhouse gas mitigation and have potential applications in biomanufacturing. Their primary metabolic enzyme, particulate methane monooxygenase ...Methane-oxidizing bacteria play a central role in greenhouse gas mitigation and have potential applications in biomanufacturing. Their primary metabolic enzyme, particulate methane monooxygenase (pMMO), is housed in copper-induced intracytoplasmic membranes (ICMs), of which the function and biogenesis are not known. We show by serial cryo-focused ion beam (cryoFIB) milling/scanning electron microscope (SEM) volume imaging and lamellae-based cellular cryo-electron tomography (cryoET) that these ICMs are derived from the inner cell membrane. The pMMO trimer, resolved by cryoET and subtomogram averaging to 4.8 Å in the ICM, forms higher-order hexagonal arrays in intact cells. Array formation correlates with increased enzymatic activity, highlighting the importance of studying the enzyme in its native environment. These findings also demonstrate the power of cryoET to structurally characterize native membrane enzymes in the cellular context. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14530.map.gz emd_14530.map.gz | 3.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14530-v30.xml emd-14530-v30.xml emd-14530.xml emd-14530.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14530.png emd_14530.png | 40.7 KB | ||
| Others |  emd_14530_half_map_1.map.gz emd_14530_half_map_1.map.gz emd_14530_half_map_2.map.gz emd_14530_half_map_2.map.gz | 1.9 MB 1.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14530 http://ftp.pdbj.org/pub/emdb/structures/EMD-14530 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14530 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14530 | HTTPS FTP |
-Validation report
| Summary document |  emd_14530_validation.pdf.gz emd_14530_validation.pdf.gz | 606.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14530_full_validation.pdf.gz emd_14530_full_validation.pdf.gz | 606.1 KB | Display | |
| Data in XML |  emd_14530_validation.xml.gz emd_14530_validation.xml.gz | 7.7 KB | Display | |
| Data in CIF |  emd_14530_validation.cif.gz emd_14530_validation.cif.gz | 8.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14530 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14530 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14530 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14530 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14530.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14530.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | pMMO trimer-trimer map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.68 Å | ||||||||||||||||||||||||||||||||||||
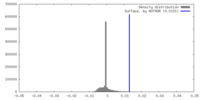
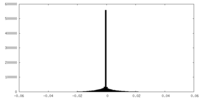
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half1 map
| File | emd_14530_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
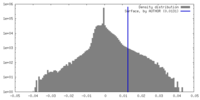
| Density Histograms |
-Half map: half2 map
| File | emd_14530_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
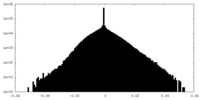
| Density Histograms |
- Sample components
Sample components
-Entire : pMMO
| Entire | Name: pMMO |
|---|---|
| Components |
|
-Supramolecule #1: pMMO
| Supramolecule | Name: pMMO / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Methylococcus capsulatus str. Bath (bacteria) Methylococcus capsulatus str. Bath (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 2 / Average electron dose: 3.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 6.0 µm / Nominal defocus min: 2.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 12.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number subtomograms used: 62250 |
|---|---|
| Extraction | Number tomograms: 187 / Number images used: 127417 |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)