+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | VP2-only capsid of MVM D263A mutant | ||||||||||||
 Map data Map data | VP2-only capsid of MVM D263A sharpened map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Capsid / MVM / VLP / VIRUS LIKE PARTICLE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont entry into host cell via permeabilization of host membrane / microtubule-dependent intracellular transport of viral material towards nucleus / T=1 icosahedral viral capsid / viral penetration into host nucleus / host cell / clathrin-dependent endocytosis of virus by host cell / host cell nucleus / virion attachment to host cell / structural molecule activity / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Minute virus of mice Minute virus of mice | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.32 Å | ||||||||||||
 Authors Authors | Luque D / Ortega-Esteban A / Valbuena A / Vilas JL / Rodriguez-Huete A / Mateu MG / Caston JR | ||||||||||||
| Funding support |  Spain, 3 items Spain, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2023 Journal: J Mol Biol / Year: 2023Title: Equilibrium Dynamics of a Biomolecular Complex Analyzed at Single-amino Acid Resolution by Cryo-electron Microscopy. Authors: Daniel Luque / Alvaro Ortega-Esteban / Alejandro Valbuena / Jose Luis Vilas / Alicia Rodríguez-Huete / Mauricio G Mateu / José R Castón /  Abstract: The biological function of macromolecular complexes depends not only on large-scale transitions between conformations, but also on small-scale conformational fluctuations at equilibrium. Information ...The biological function of macromolecular complexes depends not only on large-scale transitions between conformations, but also on small-scale conformational fluctuations at equilibrium. Information on the equilibrium dynamics of biomolecular complexes could, in principle, be obtained from local resolution (LR) data in cryo-electron microscopy (cryo-EM) maps. However, this possibility had not been validated by comparing, for a same biomolecular complex, LR data with quantitative information on equilibrium dynamics obtained by an established solution technique. In this study we determined the cryo-EM structure of the minute virus of mice (MVM) capsid as a model biomolecular complex. The LR values obtained correlated with crystallographic B factors and with hydrogen/deuterium exchange (HDX) rates obtained by mass spectrometry (HDX-MS), a gold standard for determining equilibrium dynamics in solution. This result validated a LR-based cryo-EM approach to investigate, with high spatial resolution, the equilibrium dynamics of biomolecular complexes. As an application of this approach, we determined the cryo-EM structure of two mutant MVM capsids and compared their equilibrium dynamics with that of the wild-type MVM capsid. The results supported a previously suggested linkage between mechanical stiffening and impaired equilibrium dynamics of a virus particle. Cryo-EM is emerging as a powerful approach for simultaneously acquiring information on the atomic structure and local equilibrium dynamics of biomolecular complexes. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14520.map.gz emd_14520.map.gz | 49.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14520-v30.xml emd-14520-v30.xml emd-14520.xml emd-14520.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14520.png emd_14520.png | 257.6 KB | ||
| Filedesc metadata |  emd-14520.cif.gz emd-14520.cif.gz | 5.7 KB | ||
| Others |  emd_14520_additional_1.map.gz emd_14520_additional_1.map.gz emd_14520_half_map_1.map.gz emd_14520_half_map_1.map.gz emd_14520_half_map_2.map.gz emd_14520_half_map_2.map.gz | 39.6 MB 39.4 MB 39.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14520 http://ftp.pdbj.org/pub/emdb/structures/EMD-14520 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14520 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14520 | HTTPS FTP |
-Validation report
| Summary document |  emd_14520_validation.pdf.gz emd_14520_validation.pdf.gz | 816.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14520_full_validation.pdf.gz emd_14520_full_validation.pdf.gz | 815.8 KB | Display | |
| Data in XML |  emd_14520_validation.xml.gz emd_14520_validation.xml.gz | 11.7 KB | Display | |
| Data in CIF |  emd_14520_validation.cif.gz emd_14520_validation.cif.gz | 13.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14520 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14520 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14520 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14520 | HTTPS FTP |
-Related structure data
| Related structure data |  7z5eMC  7z5dC  7z5fC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14520.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14520.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | VP2-only capsid of MVM D263A sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.37 Å | ||||||||||||||||||||||||||||||||||||
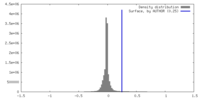
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: VP2-only capsid of MVM D263A unsharpened map
| File | emd_14520_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | VP2-only capsid of MVM D263A unsharpened map | ||||||||||||
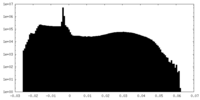
| Projections & Slices |
| ||||||||||||
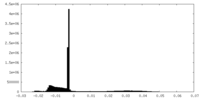
| Density Histograms |
-Half map: VP2-only capsid of MVM D263A half map 2
| File | emd_14520_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | VP2-only capsid of MVM D263A half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: VP2-only capsid of MVM D263A half map 1
| File | emd_14520_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | VP2-only capsid of MVM D263A half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Minute virus of mice
| Entire | Name:  Minute virus of mice Minute virus of mice |
|---|---|
| Components |
|
-Supramolecule #1: Minute virus of mice
| Supramolecule | Name: Minute virus of mice / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10794 / Sci species name: Minute virus of mice / Sci species strain: prototype / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Minute virus of mice / Strain: MVM prototype Minute virus of mice / Strain: MVM prototype |
| Molecular weight | Theoretical: 64.558461 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSDGTSQPDS GNAVHSAARV ERAADGPGGS GGGGSGGGGV GVSTGSYDNQ THYRFLGDGW VEITALATRL VHLNMPKSEN YCRIRVHNT TDTSVKGNMA KDDAHEQIWT PWSLVDANAW GVWLQPSDWQ YICNTMSQLN LVSLDQEIFN VVLKTVTEQD L GGQAIKIY ...String: MSDGTSQPDS GNAVHSAARV ERAADGPGGS GGGGSGGGGV GVSTGSYDNQ THYRFLGDGW VEITALATRL VHLNMPKSEN YCRIRVHNT TDTSVKGNMA KDDAHEQIWT PWSLVDANAW GVWLQPSDWQ YICNTMSQLN LVSLDQEIFN VVLKTVTEQD L GGQAIKIY NNDLTACMMV AVDSNNILPY TPAANSMETL GFYPWKPTIA SPYRYYFCVD RDLSVTYENQ EGTVEHNVMG TP KGMNSQF FTIENTQQIT LLRTGAEFAT GTYYFDTNPV KLTHTWQTNR QLGQPPLLST FPEADTDAGT LTAQGSRHGT TQM GVNWVS EAIRTRPAQV GFCQPHNDFE ASRAGPFAAP KVPADITQGV DKEANGSVRY SYGKQHGENW ASHGPAPERY TWDE TSFGS GRDTKDGFIQ SAPLVVPPPL NGILTNANPI GTKNDIHFSN VFNSYGPLTA FSHPSPVYPQ GQIWDKELDL EHKPR LHIT APFVCKNNAP GQMLVRLGPN LTDQYDPNGA TLSRIVTYGT FFWKGKLTMR AKLRANTTWN PVYQVSAEDN GNSYMS VTK WLPTATGNMQ SVPLITRPVA RNTY UniProtKB: Capsid protein VP1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM CPC |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 36.6 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)