[English] 日本語
 Yorodumi
Yorodumi- EMDB-14511: Human NEXT dimer - focused reconstruction of the dimerization module -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human NEXT dimer - focused reconstruction of the dimerization module | |||||||||||||||
 Map data Map data | NEXT_S_dimerization_module_postprocess | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | HELICASE / ATPASE / RNA DEGRADATION / EXOSOME / RNA BINDING PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsnRNA catabolic process / TRAMP complex / mRNA 3'-end processing / RNA catabolic process / maturation of 5.8S rRNA / Major pathway of rRNA processing in the nucleolus and cytosol / RNA processing / catalytic step 2 spliceosome / mRNA Splicing - Major Pathway / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex ...snRNA catabolic process / TRAMP complex / mRNA 3'-end processing / RNA catabolic process / maturation of 5.8S rRNA / Major pathway of rRNA processing in the nucleolus and cytosol / RNA processing / catalytic step 2 spliceosome / mRNA Splicing - Major Pathway / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / mRNA splicing, via spliceosome / rRNA processing / RNA helicase activity / nuclear body / RNA helicase / nuclear speck / DNA damage response / nucleolus / ATP hydrolysis activity / RNA binding / zinc ion binding / nucleoplasm / ATP binding / nucleus Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||||||||
 Authors Authors | Gerlach P / Lingaraju M / Salerno-Kochan A / Bonneau F / Basquin J / Conti E | |||||||||||||||
| Funding support |  Germany, European Union, 4 items Germany, European Union, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome. Authors: Piotr Gerlach / William Garland / Mahesh Lingaraju / Anna Salerno-Kochan / Fabien Bonneau / Jérôme Basquin / Torben Heick Jensen / Elena Conti /   Abstract: In mammalian cells, spurious transcription results in a vast repertoire of unproductive non-coding RNAs, whose deleterious accumulation is prevented by rapid decay. The nuclear exosome targeting ...In mammalian cells, spurious transcription results in a vast repertoire of unproductive non-coding RNAs, whose deleterious accumulation is prevented by rapid decay. The nuclear exosome targeting (NEXT) complex plays a central role in directing non-functional transcripts to exosome-mediated degradation, but the structural and molecular mechanisms remain enigmatic. Here, we elucidated the architecture of the human NEXT complex, showing that it exists as a dimer of MTR4-ZCCHC8-RBM7 heterotrimers. Dimerization preconfigures the major MTR4-binding region of ZCCHC8 and arranges the two MTR4 helicases opposite to each other, with each protomer able to function on many types of RNAs. In the inactive state of the complex, the 3' end of an RNA substrate is enclosed in the MTR4 helicase channel by a ZCCHC8 C-terminal gatekeeping domain. The architecture of a NEXT-exosome assembly points to the molecular and regulatory mechanisms with which the NEXT complex guides RNA substrates to the exosome. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14511.map.gz emd_14511.map.gz | 28.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14511-v30.xml emd-14511-v30.xml emd-14511.xml emd-14511.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14511_fsc.xml emd_14511_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_14511.png emd_14511.png | 113.3 KB | ||
| Masks |  emd_14511_msk_1.map emd_14511_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14511.cif.gz emd-14511.cif.gz | 6.3 KB | ||
| Others |  emd_14511_additional_1.map.gz emd_14511_additional_1.map.gz emd_14511_half_map_1.map.gz emd_14511_half_map_1.map.gz emd_14511_half_map_2.map.gz emd_14511_half_map_2.map.gz | 1.4 MB 23.5 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14511 http://ftp.pdbj.org/pub/emdb/structures/EMD-14511 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14511 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14511 | HTTPS FTP |
-Validation report
| Summary document |  emd_14511_validation.pdf.gz emd_14511_validation.pdf.gz | 935.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14511_full_validation.pdf.gz emd_14511_full_validation.pdf.gz | 935.2 KB | Display | |
| Data in XML |  emd_14511_validation.xml.gz emd_14511_validation.xml.gz | 12.4 KB | Display | |
| Data in CIF |  emd_14511_validation.cif.gz emd_14511_validation.cif.gz | 17.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14511 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14511 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14511 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14511 | HTTPS FTP |
-Related structure data
| Related structure data |  7z4zMC  7z4yC  7z52C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14511.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14511.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | NEXT_S_dimerization_module_postprocess | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.094 Å | ||||||||||||||||||||||||||||||||||||
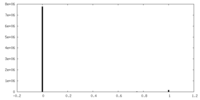
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14511_msk_1.map emd_14511_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
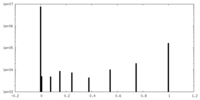
| Density Histograms |
-Additional map: NEXT S dimerization module refine 238 denmod map
| File | emd_14511_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | NEXT_S_dimerization_module_refine_238_denmod_map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: NEXT S dimerization module half map 2
| File | emd_14511_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | NEXT_S_dimerization_module_half_map_2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: NEXT S dimerization module half map 1
| File | emd_14511_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | NEXT_S_dimerization_module_half_map_1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human NEXT dimer - focused reconstruction of the dimerization module
| Entire | Name: Human NEXT dimer - focused reconstruction of the dimerization module |
|---|---|
| Components |
|
-Supramolecule #1: Human NEXT dimer - focused reconstruction of the dimerization module
| Supramolecule | Name: Human NEXT dimer - focused reconstruction of the dimerization module type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Zinc finger CCHC domain-containing protein 8
| Macromolecule | Name: Zinc finger CCHC domain-containing protein 8 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.668961 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPDSENGVGD AELRERLRQC EETIEQLRAE NQELKRKLNI LTRPSGILVN DTKLDGPILQ ILFMNNAISK QYHQEIEEFV SNLVKRFEE QQKNDVEKTS FNLLPQPSSI VLEEDHKVEE SCAIKNNKEA FSVVGSVLYF TNFCLDKLGQ PLLNENPQLS E GWEIPKYH ...String: GPDSENGVGD AELRERLRQC EETIEQLRAE NQELKRKLNI LTRPSGILVN DTKLDGPILQ ILFMNNAISK QYHQEIEEFV SNLVKRFEE QQKNDVEKTS FNLLPQPSSI VLEEDHKVEE SCAIKNNKEA FSVVGSVLYF TNFCLDKLGQ PLLNENPQLS E GWEIPKYH QVFSHIVSLE GQEIQVKAKR PKPHCFNCGS EEHQMKDCPM PRNAARISEK RKEYMDACGE ANNQNFQQRY HA EEVEERF GRFKPGVISE ELQDALGVTD KSLPPFIYRM RQLGYPPGWL KEAELENSGL ALYD UniProtKB: Zinc finger CCHC domain-containing protein 8 |
-Macromolecule #2: Exosome RNA helicase MTR4
| Macromolecule | Name: Exosome RNA helicase MTR4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: RNA helicase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 118.32207 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPDSMADAFG DELFSVFEGD STTAAGTKKD KEKDKGKWKG PPGSADKAGK RFDGKLQSES TNNGKNKRDV DFEGTDEPIF GKKPRIEES ITEDLSLADL MPRVKVQSVE TVEGCTHEVA LPAEEDYLPL KPRVGKAAKE YPFILDAFQR EAIQCVDNNQ S VLVSAHTS ...String: GPDSMADAFG DELFSVFEGD STTAAGTKKD KEKDKGKWKG PPGSADKAGK RFDGKLQSES TNNGKNKRDV DFEGTDEPIF GKKPRIEES ITEDLSLADL MPRVKVQSVE TVEGCTHEVA LPAEEDYLPL KPRVGKAAKE YPFILDAFQR EAIQCVDNNQ S VLVSAHTS AGKTVCAEYA IALALREKQR VIFTSPIKAL SNQKYREMYE EFQDVGLMTG DVTINPTASC LVMTTEILRS ML YRGSEVM REVAWVIFDE IHYMRDSERG VVWEETIILL PDNVHYVFLS ATIPNARQFA EWICHLHKQP CHVIYTDYRP TPL QHYIFP AGGDGLHLVV DENGDFREDN FNTAMQVLRD AGDLAKGDQK GRKGGTKGPS NVFKIVKMIM ERNFQPVIIF SFSK KDCEA YALQMTKLDF NTDEEKKMVE EVFSNAIDCL SDEDKKLPQV EHVLPLLKRG IGIHHGGLLP ILKETIEILF SEGLI KALF ATETFAMGIN MPARTVLFTN ARKFDGKDFR WISSGEYIQM SGRAGRRGMD DRGIVILMVD EKMSPTIGKQ LLKGSA DPL NSAFHLTYNM VLNLLRVEEI NPEYMLEKSF YQFQHYRAIP GVVEKVKNSE EQYNKIVIPN EESVVIYYKI RQQLAKL GK EIEEYIHKPK YCLPFLQPGR LVKVKNEGDD FGWGVVVNFS KKSNVKPNSG ELDPLYVVEV LLRCSKESLK NSATEAAK P AKPDEKGEMQ VVPVLVHLLS AISSVRLYIP KDLRPVDNRQ SVLKSIQEVQ KRFPDGIPLL DPIDDMGIQD QGLKKVIQK VEAFEHRMYS HPLHNDPNLE TVYTLCEKKA QIAIDIKSAK RELKKARTVL QMDELKCRKR VLRRLGFATS SDVIEMKGRV ACEISSADE LLLTEMMFNG LFNDLSAEQA TALLSCFVFQ ENSSEMPKLT EQLAGPLRQM QECAKRIAKV SAEAKLEIDE E TYLSSFKP HLMDVVYTWA TGATFAHICK MTDVFEGSII RCMRRLEELL RQMCQAAKAI GNTELENKFA EGITKIKRDI VF AASLYL UniProtKB: Exosome RNA helicase MTR4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 2.09 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)