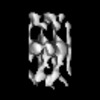[English] 日本語
 Yorodumi
Yorodumi- EMDB-1449: The molecular architecture of cadherins in native epidermal desmo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1449 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The molecular architecture of cadherins in native epidermal desmosomes. | |||||||||
 Map data Map data | Image used for the isosurface representation, after segmentation and mild filtering | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 34.0 Å | |||||||||
 Authors Authors | Al-Amoudi A / Diez DC / Betts MJ / Frangakis AS | |||||||||
 Citation Citation |  Journal: Nature / Year: 2007 Journal: Nature / Year: 2007Title: The molecular architecture of cadherins in native epidermal desmosomes. Authors: Ashraf Al-Amoudi / Daniel Castaño Díez / Matthew J Betts / Achilleas S Frangakis /  Abstract: Desmosomes are cadherin-based adhesive intercellular junctions, which are present in tissues such as heart and skin. Despite considerable efforts, the molecular interfaces that mediate adhesion ...Desmosomes are cadherin-based adhesive intercellular junctions, which are present in tissues such as heart and skin. Despite considerable efforts, the molecular interfaces that mediate adhesion remain obscure. Here we apply cryo-electron tomography of vitreous sections from human epidermis to visualize the three-dimensional molecular architecture of desmosomal cadherins at close-to-native conditions. The three-dimensional reconstructions show a regular array of densities at approximately 70 A intervals along the midline, with a curved shape resembling the X-ray structure of C-cadherin, a representative 'classical' cadherin. Model-independent three-dimensional image processing of extracted sub-tomograms reveals the cadherin organization. After fitting the C-cadherin atomic structure into the averaged sub-tomograms, we see a periodic arrangement of a trans W-like and a cis V-like interaction corresponding to molecules from opposing membranes and the same cell membrane, respectively. The resulting model of cadherin organization explains existing two-dimensional data and yields insights into a possible mechanism of cadherin-based cell adhesion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1449.map.gz emd_1449.map.gz | 728.2 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1449-v30.xml emd-1449-v30.xml emd-1449.xml emd-1449.xml | 8.4 KB 8.4 KB | Display Display |  EMDB header EMDB header |
| Images |  1449.gif 1449.gif | 11 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1449 http://ftp.pdbj.org/pub/emdb/structures/EMD-1449 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1449 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1449 | HTTPS FTP |
-Validation report
| Summary document |  emd_1449_validation.pdf.gz emd_1449_validation.pdf.gz | 202.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1449_full_validation.pdf.gz emd_1449_full_validation.pdf.gz | 201.7 KB | Display | |
| Data in XML |  emd_1449_validation.xml.gz emd_1449_validation.xml.gz | 4.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1449 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1449 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1449 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1449 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1449.map.gz / Format: CCP4 / Size: 1.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1449.map.gz / Format: CCP4 / Size: 1.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Image used for the isosurface representation, after segmentation and mild filtering | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
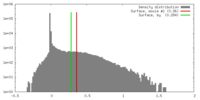
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cadherin Organisation in Native Desmosomes
| Entire | Name: Cadherin Organisation in Native Desmosomes |
|---|---|
| Components |
|
-Supramolecule #1000: Cadherin Organisation in Native Desmosomes
| Supramolecule | Name: Cadherin Organisation in Native Desmosomes / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Desmosome
| Supramolecule | Name: Desmosome / type: organelle_or_cellular_component / ID: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Tissue: Epidermis Homo sapiens (human) / synonym: Human / Tissue: Epidermis |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Grid | Details: 200 mesh quantifoil grid |
|---|---|
| Vitrification | Cryogen name: NITROGEN / Instrument: OTHER / Details: Vitrification instrument: Leica EMPact 2 / Method: High pressure freezing |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Temperature | Average: 100 K |
| Specialist optics | Energy filter - Name: Gatan 2002 / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 30.0 eV |
| Image recording | Average electron dose: 40 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 49669 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus min: 3.8 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN / Tilt series - Axis1 - Min angle: 64.0 ° / Tilt series - Axis1 - Max angle: 64 ° |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Average number of projections used in the 3D reconstructions: 417. |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 34.0 Å / Resolution method: FSC 0.5 CUT-OFF |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Details | Protocol: Rigid body. Semi-automated fitting |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)