[English] 日本語
 Yorodumi
Yorodumi- EMDB-14393: Hairpin-bound state of the E. coli Mre11-Rad50 (SbcCD) head compl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hairpin-bound state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and a DNA hairpin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC-type ATPase / Nuclease / DNA repair / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationdouble-stranded DNA endonuclease activity / DNA replication termination / DNA exonuclease activity / single-stranded DNA endodeoxyribonuclease activity / 3'-5'-DNA exonuclease activity / DNA repair complex / 3'-5' exonuclease activity / double-strand break repair / DNA recombination / DNA replication ...double-stranded DNA endonuclease activity / DNA replication termination / DNA exonuclease activity / single-stranded DNA endodeoxyribonuclease activity / 3'-5'-DNA exonuclease activity / DNA repair complex / 3'-5' exonuclease activity / double-strand break repair / DNA recombination / DNA replication / DNA repair / ATP hydrolysis activity / DNA binding / ATP binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
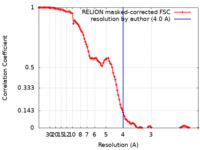
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Gut F / Kaeshammer L | |||||||||
| Funding support | European Union,  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50. Authors: Fabian Gut / Lisa Käshammer / Katja Lammens / Joseph D Bartho / Anna-Maria Boggusch / Erik van de Logt / Brigitte Kessler / Karl-Peter Hopfner /  Abstract: DNA double-strand breaks (DSBs) threaten genome stability and are linked to tumorigenesis in humans. Repair of DSBs requires the removal of attached proteins and hairpins through a poorly understood ...DNA double-strand breaks (DSBs) threaten genome stability and are linked to tumorigenesis in humans. Repair of DSBs requires the removal of attached proteins and hairpins through a poorly understood but physiologically critical endonuclease activity by the Mre11-Rad50 complex. Here, we report cryoelectron microscopy (cryo-EM) structures of the bacterial Mre11-Rad50 homolog SbcCD bound to a protein-blocked DNA end and a DNA hairpin. The structures reveal that Mre11-Rad50 bends internal DNA for endonucleolytic cleavage and show how internal DNA, DNA ends, and hairpins are processed through a similar ATP-regulated conformational state. Furthermore, Mre11-Rad50 is loaded onto blocked DNA ends with Mre11 pointing away from the block, explaining the distinct biochemistries of 3' → 5' exonucleolytic and endonucleolytic incision through the way Mre11-Rad50 interacts with diverse DNA ends. In summary, our results unify Mre11-Rad50's enigmatic nuclease diversity within a single structural framework and reveal how blocked DNA ends and hairpins are processed. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14393.map.gz emd_14393.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14393-v30.xml emd-14393-v30.xml emd-14393.xml emd-14393.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14393_fsc.xml emd_14393_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_14393.png emd_14393.png | 71.9 KB | ||
| Masks |  emd_14393_msk_1.map emd_14393_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14393.cif.gz emd-14393.cif.gz | 6.8 KB | ||
| Others |  emd_14393_additional_1.map.gz emd_14393_additional_1.map.gz emd_14393_half_map_1.map.gz emd_14393_half_map_1.map.gz emd_14393_half_map_2.map.gz emd_14393_half_map_2.map.gz | 951.9 MB 98.2 MB 98.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14393 http://ftp.pdbj.org/pub/emdb/structures/EMD-14393 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14393 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14393 | HTTPS FTP |
-Validation report
| Summary document |  emd_14393_validation.pdf.gz emd_14393_validation.pdf.gz | 799 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14393_full_validation.pdf.gz emd_14393_full_validation.pdf.gz | 798.6 KB | Display | |
| Data in XML |  emd_14393_validation.xml.gz emd_14393_validation.xml.gz | 18.4 KB | Display | |
| Data in CIF |  emd_14393_validation.cif.gz emd_14393_validation.cif.gz | 24.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14393 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14393 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14393 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14393 | HTTPS FTP |
-Related structure data
| Related structure data |  7yzpMC  7yzoC  7z03C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14393.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14393.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||
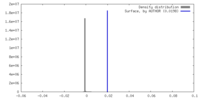
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14393_msk_1.map emd_14393_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
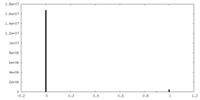
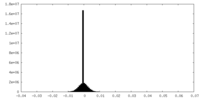
| Density Histograms |
-Additional map: LAFTER-filtered map
| File | emd_14393_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LAFTER-filtered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
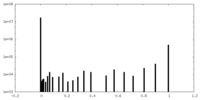
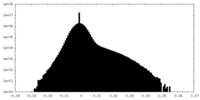
| Density Histograms |
-Half map: #2
| File | emd_14393_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
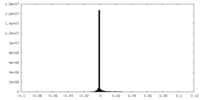
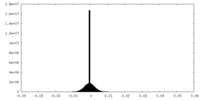
| Density Histograms |
-Half map: #1
| File | emd_14393_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
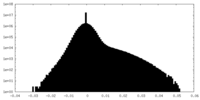
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of Mre11 dimer, Rad50 dimer and a DNA hairpin
| Entire | Name: Complex of Mre11 dimer, Rad50 dimer and a DNA hairpin |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Mre11 dimer, Rad50 dimer and a DNA hairpin
| Supramolecule | Name: Complex of Mre11 dimer, Rad50 dimer and a DNA hairpin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: Nuclease SbcCD
| Supramolecule | Name: Nuclease SbcCD / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: DNA hairpin
| Supramolecule | Name: DNA hairpin / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
-Macromolecule #1: DNA hairpin (59-MER)
| Macromolecule | Name: DNA hairpin (59-MER) / type: dna / ID: 1 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 23.144822 KDa |
| Sequence | String: (DC)(DG)(DC)(DT)(DT)(DT)(DA)(DT)(DC)(DA) (DG)(DA)(DA)(DG)(DC)(DC)(DA)(DG)(DA)(DC) (DA)(DT)(DT)(DA)(DA)(DC)(DG)(DC)(DT) (DT)(DC)(DT)(DG)(DG)(DA)(DT)(DA)(DG)(DC) (DG) (DT)(DC)(DC)(DA)(DG)(DA) ...String: (DC)(DG)(DC)(DT)(DT)(DT)(DA)(DT)(DC)(DA) (DG)(DA)(DA)(DG)(DC)(DC)(DA)(DG)(DA)(DC) (DA)(DT)(DT)(DA)(DA)(DC)(DG)(DC)(DT) (DT)(DC)(DT)(DG)(DG)(DA)(DT)(DA)(DG)(DC) (DG) (DT)(DC)(DC)(DA)(DG)(DA)(DA)(DG) (DC)(DG)(DT)(DT)(DA)(DA)(DT)(DG)(DT)(DC) (DT)(DG) (DG)(DC)(DT)(DT)(DC)(DT)(DG) (DA)(DT)(DA)(DA)(DA)(DG)(DC)(DG) |
-Macromolecule #2: Nuclease SbcCD subunit D
| Macromolecule | Name: Nuclease SbcCD subunit D / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.640277 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRILHTSDWH LGQNFYSKSR EAEHQAFLDW LLETAQTHQV DAIIVAGDVF DTGSPPSYAR TLYNRFVVNL QQTGCHLVVL AGNQDSVAT LNESRDIMAF LNTTVVASAG HAPQILPRRD GTPGAVLCPI PFLRPRDIIT SQAGLNGIEK QQHLLAAITD Y YQQHYADA ...String: MRILHTSDWH LGQNFYSKSR EAEHQAFLDW LLETAQTHQV DAIIVAGDVF DTGSPPSYAR TLYNRFVVNL QQTGCHLVVL AGNQDSVAT LNESRDIMAF LNTTVVASAG HAPQILPRRD GTPGAVLCPI PFLRPRDIIT SQAGLNGIEK QQHLLAAITD Y YQQHYADA CKLRGDQPLP IIATGHLTTV GASKSDAVRD IYIGTLDAFP AQNFPPADYI ALGHIHRAQI IGGMEHVRYC GS PIPLSFD ECGKSKYVHL VTFSNGKLES VENLNVPVTQ PMAVLKGDLA SITAQLEQWR DVSQEPPVWL DIEITTDEYL HDI QRKIQA LTESLPVEVL LVRRSREQRE RVLASQQRET LSELSVEEVF NRRLALEELD ESQQQRLQHL FTTTLHTLAG EHEA GHHHH HH UniProtKB: Nuclease SbcCD subunit D |
-Macromolecule #3: Nuclease SbcCD subunit C
| Macromolecule | Name: Nuclease SbcCD subunit C / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 118.851508 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKILSLRLKN LNSLKGEWKI DFTREPFASN GLFAITGPTG AGKTTLLDAI CLALYHETPR LSNVSQSQND LMTRDTAECL AEVEFEVKG EAYRAFWSQN RARNQPDGNL QVPRVELARC ADGKILADKV KDKLELTATL TGLDYGRFTR SMLLSQGQFA A FLNAKPKE ...String: MKILSLRLKN LNSLKGEWKI DFTREPFASN GLFAITGPTG AGKTTLLDAI CLALYHETPR LSNVSQSQND LMTRDTAECL AEVEFEVKG EAYRAFWSQN RARNQPDGNL QVPRVELARC ADGKILADKV KDKLELTATL TGLDYGRFTR SMLLSQGQFA A FLNAKPKE RAELLEELTG TEIYGQISAM VFEQHKSART ELEKLQAQAS GVTLLTPEQV QSLTASLQVL TDEEKQLITA QQ QEQQSLN WLTRQDELQQ EASRRQQALQ QALAEEEKAQ PQLAALSLAQ PARNLRPHWE RIAEHSAALA HIRQQIEEVN TRL QSTMAL RASIRHHAAK QSAELQQQQQ SLNTWLQEHD RFRQWNNEPA GWRAQFSQQT SDREHLRQWQ QQLTHAEQKL NALA AITLT LTADEVATAL AQHAEQRPLR QHLVALHGQI VPQQKRLAQL QVAIQNVTQE QTQRNAALNE MRQRYKEKTQ QLADV KTIC EQEARIKTLE AQRAQLQAGQ PCPLCGSTSH PAVEAYQALE PGVNQSRLLA LENEVKKLGE EGATLRGQLD AITKQL QRD ENEAQSLRQD EQALTQQWQA VTASLNITLQ PLDDIQPWLD AQDEHERQLR LLSQRHELQG QIAAHNQQII QYQQQIE QR QQLLLTTLTG YALTLPQEDE EESWLATRQQ EAQSWQQRQN ELTALQNRIQ QLTPILETLP QSDELPHCEE TVVLENWR Q VHEQCLALHS QQQTLQQQDV LAAQSLQKAQ AQFDTALQAS VFDDQQAFLA ALMDEQTLTQ LEQLKQNLEN QRRQAQTLV TQTAETLAQH QQHRPDDGLA LTVTVEQIQQ ELAQTHQKLR ENTTSQGEIR QQLKQDADNR QQQQTLMQQI AQMTQQVEDW GYLNSLIGS KEGDKFRKFA QGLTLDNLVH LANQQLTRLH GRYLLQRKAS EALEVEVVDT WQADAVRDTR TLSGGESFLV S LALALALS DLVSHKTRID SLFLDEGFGT LDSETLDTAL DALDALNASG KTIGVISHVE AMKERIPVQI KVKKINGLGY SK LESTFAV K UniProtKB: Nuclease SbcCD subunit C |
-Macromolecule #4: MANGANESE (II) ION
| Macromolecule | Name: MANGANESE (II) ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MN |
|---|---|
| Molecular weight | Theoretical: 54.938 Da |
-Macromolecule #5: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 46.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)