+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of Bacillus megaterium gas vesicles | |||||||||
 マップデータ マップデータ | Main map, automatically B-factor sharpened with -30 A**2 in cryoSPARC / symmetrized with -3.874 degree rotation, 0.525 A rise | |||||||||
 試料 試料 |
| |||||||||
| 生物種 |  Priestia megaterium NBRC 15308 = ATCC 14581 (バクテリア) Priestia megaterium NBRC 15308 = ATCC 14581 (バクテリア) | |||||||||
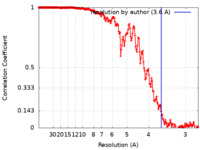
| 手法 | らせん対称体再構成法 / クライオ電子顕微鏡法 / 解像度: 3.6 Å | |||||||||
 データ登録者 データ登録者 | Huber ST / Evers W / Jakobi AJ | |||||||||
| 資金援助 | 1件
| |||||||||
 引用 引用 |  ジャーナル: Cell / 年: 2023 ジャーナル: Cell / 年: 2023タイトル: Cryo-EM structure of gas vesicles for buoyancy-controlled motility. 著者: Stefan T Huber / Dion Terwiel / Wiel H Evers / David Maresca / Arjen J Jakobi /  要旨: Gas vesicles are gas-filled nanocompartments that allow a diverse group of bacteria and archaea to control their buoyancy. The molecular basis of their properties and assembly remains unclear. Here, ...Gas vesicles are gas-filled nanocompartments that allow a diverse group of bacteria and archaea to control their buoyancy. The molecular basis of their properties and assembly remains unclear. Here, we report the 3.2 Å cryo-EM structure of the gas vesicle shell made from the structural protein GvpA that self-assembles into hollow helical cylinders closed off by cone-shaped tips. Two helical half shells connect through a characteristic arrangement of GvpA monomers, suggesting a mechanism of gas vesicle biogenesis. The fold of GvpA features a corrugated wall structure typical for force-bearing thin-walled cylinders. Small pores enable gas molecules to diffuse across the shell, while the exceptionally hydrophobic interior surface effectively repels water. Comparative structural analysis confirms the evolutionary conservation of gas vesicle assemblies and demonstrates molecular features of shell reinforcement by GvpC. Our findings will further research into gas vesicle biology and facilitate molecular engineering of gas vesicles for ultrasound imaging. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_14340.map.gz emd_14340.map.gz | 115.6 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-14340-v30.xml emd-14340-v30.xml emd-14340.xml emd-14340.xml | 19.5 KB 19.5 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_14340_fsc.xml emd_14340_fsc.xml | 17.7 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_14340.png emd_14340.png | 245.5 KB | ||
| マスクデータ |  emd_14340_msk_1.map emd_14340_msk_1.map | 512 MB |  マスクマップ マスクマップ | |
| その他 |  emd_14340_additional_1.map.gz emd_14340_additional_1.map.gz emd_14340_half_map_1.map.gz emd_14340_half_map_1.map.gz emd_14340_half_map_2.map.gz emd_14340_half_map_2.map.gz | 115.5 MB 475.4 MB 475.5 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14340 http://ftp.pdbj.org/pub/emdb/structures/EMD-14340 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14340 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14340 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_14340_validation.pdf.gz emd_14340_validation.pdf.gz | 892.9 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_14340_full_validation.pdf.gz emd_14340_full_validation.pdf.gz | 892.5 KB | 表示 | |
| XML形式データ |  emd_14340_validation.xml.gz emd_14340_validation.xml.gz | 26.4 KB | 表示 | |
| CIF形式データ |  emd_14340_validation.cif.gz emd_14340_validation.cif.gz | 34.3 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14340 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14340 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14340 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14340 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_14340.map.gz / 形式: CCP4 / 大きさ: 512 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_14340.map.gz / 形式: CCP4 / 大きさ: 512 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Main map, automatically B-factor sharpened with -30 A**2 in cryoSPARC / symmetrized with -3.874 degree rotation, 0.525 A rise | ||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.37 Å | ||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-マスク #1
| ファイル |  emd_14340_msk_1.map emd_14340_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
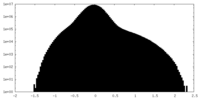
| 投影像・断面図 |
| ||||||||||||
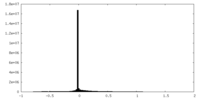
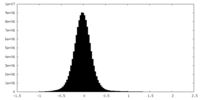
| 密度ヒストグラム |
-追加マップ: Unsharpened map / symmetrized with -3.874 degree rotation,...
| ファイル | emd_14340_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Unsharpened map / symmetrized with -3.874 degree rotation, 0.525 A rise | ||||||||||||
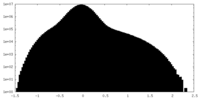
| 投影像・断面図 |
| ||||||||||||
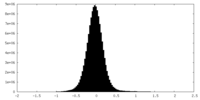
| 密度ヒストグラム |
-ハーフマップ: Half Map A
| ファイル | emd_14340_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half Map A | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Half Map B
| ファイル | emd_14340_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half Map B | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Helical assembly of GvpB monomers forming the gas vesicle wall
| 全体 | 名称: Helical assembly of GvpB monomers forming the gas vesicle wall |
|---|---|
| 要素 |
|
-超分子 #1: Helical assembly of GvpB monomers forming the gas vesicle wall
| 超分子 | 名称: Helical assembly of GvpB monomers forming the gas vesicle wall タイプ: complex / ID: 1 / キメラ: Yes / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  Priestia megaterium NBRC 15308 = ATCC 14581 (バクテリア) Priestia megaterium NBRC 15308 = ATCC 14581 (バクテリア) |
| 分子量 | 理論値: 9.99 MDa |
-分子 #1: GvpA2/B from B.megaterium
| 分子 | 名称: GvpA2/B from B.megaterium / タイプ: protein_or_peptide / ID: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Priestia megaterium NBRC 15308 = ATCC 14581 (バクテリア) Priestia megaterium NBRC 15308 = ATCC 14581 (バクテリア) |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MSIQKSTNSS SLAEVIDRI L DKGIVIDA FA RVSVVGI EIL TIEARV VIAS VDTWL RYAEA VGLL RDDVEE NGL PERSNSS EG QPRFSI |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | らせん対称体再構成法 |
| 試料の集合状態 | helical array |
- 試料調製
試料調製
| 濃度 | 0.45 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 8 構成要素:
| |||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 95 % / チャンバー内温度: 293 K / 装置: LEICA PLUNGER / 詳細: Blot times between 5 and 11 seconds.. | |||||||||
| 詳細 | Concentration measured by OD(500)=3.12 against a sonicated blank. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) デジタル化 - サイズ - 横: 5760 pixel / デジタル化 - サイズ - 縦: 4092 pixel / 撮影したグリッド数: 1 / 実像数: 4351 / 平均露光時間: 2.4 sec. / 平均電子線量: 30.0 e/Å2 詳細: One shot per hole 1.37 A/pix 60 fractions over 30 e-/A2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 1.25 µm / 最小 デフォーカス(公称値): 0.25 µm / 倍率(公称値): 64000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 精密化 | 空間: REAL / プロトコル: AB INITIO MODEL |
|---|
 ムービー
ムービー コントローラー
コントローラー






 Z
Z Y
Y X
X