[English] 日本語
 Yorodumi
Yorodumi- EMDB-14314: The SARS-CoV-2 spike in complex with the 1.29 neutralizing nanobody -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The SARS-CoV-2 spike in complex with the 1.29 neutralizing nanobody | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | coronavirus / COVID-19 / SARS-CoV-2 / spike / nanobodies / neutralization / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
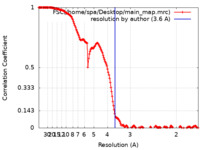
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Casasnovas JM / Melero R | |||||||||
| Funding support |  Spain, European Union, 2 items Spain, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Front Immunol / Year: 2022 Journal: Front Immunol / Year: 2022Title: Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron. Authors: José M Casasnovas / Yago Margolles / María A Noriega / María Guzmán / Rocío Arranz / Roberto Melero / Mercedes Casanova / Juan Alberto Corbera / Nereida Jiménez-de-Oya / Pablo ...Authors: José M Casasnovas / Yago Margolles / María A Noriega / María Guzmán / Rocío Arranz / Roberto Melero / Mercedes Casanova / Juan Alberto Corbera / Nereida Jiménez-de-Oya / Pablo Gastaminza / Urtzi Garaigorta / Juan Carlos Saiz / Miguel Ángel Martín-Acebes / Luis Ángel Fernández /  Abstract: The emergence of SARS-CoV-2 variants that escape from immune neutralization are challenging vaccines and antibodies developed to stop the COVID-19 pandemic. Thus, it is important to establish ...The emergence of SARS-CoV-2 variants that escape from immune neutralization are challenging vaccines and antibodies developed to stop the COVID-19 pandemic. Thus, it is important to establish therapeutics directed toward multiple or specific SARS-CoV-2 variants. The envelope spike (S) glycoprotein of SARS-CoV-2 is the key target of neutralizing antibodies (Abs). We selected a panel of nine nanobodies (Nbs) from dromedary camels immunized with the receptor-binding domain (RBD) of the S, and engineered Nb fusions as humanized heavy chain Abs (hcAbs). Nbs and derived hcAbs bound with subnanomolar or picomolar affinities to the S and its RBD, and S-binding cross-competition clustered them in two different groups. Most of the hcAbs hindered RBD binding to its human ACE2 (hACE2) receptor, blocked cell entry of viruses pseudotyped with the S protein and neutralized SARS-CoV-2 infection in cell cultures. Four potent neutralizing hcAbs prevented the progression to lethal SARS-CoV-2 infection in hACE2-transgenic mice, demonstrating their therapeutic potential. Cryo-electron microscopy identified Nb binding epitopes in and out the receptor binding motif (RBM), and showed different ways to prevent virus binding to its cell entry receptor. The Nb binding modes were consistent with its recognition of SARS-CoV-2 RBD variants; mono and bispecific hcAbs efficiently bound all variants of concern except omicron, which emphasized the immune escape capacity of this latest variant. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14314.map.gz emd_14314.map.gz | 404 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14314-v30.xml emd-14314-v30.xml emd-14314.xml emd-14314.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14314_fsc.xml emd_14314_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_14314.png emd_14314.png | 47.8 KB | ||
| Filedesc metadata |  emd-14314.cif.gz emd-14314.cif.gz | 7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14314 http://ftp.pdbj.org/pub/emdb/structures/EMD-14314 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14314 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14314 | HTTPS FTP |
-Validation report
| Summary document |  emd_14314_validation.pdf.gz emd_14314_validation.pdf.gz | 352.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14314_full_validation.pdf.gz emd_14314_full_validation.pdf.gz | 351.7 KB | Display | |
| Data in XML |  emd_14314_validation.xml.gz emd_14314_validation.xml.gz | 15.9 KB | Display | |
| Data in CIF |  emd_14314_validation.cif.gz emd_14314_validation.cif.gz | 21.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14314 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14314 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14314 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14314 | HTTPS FTP |
-Related structure data
| Related structure data |  7r4qMC  7r4iC  7r4rC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14314.map.gz / Format: CCP4 / Size: 454.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14314.map.gz / Format: CCP4 / Size: 454.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : The trimeric SARS-CooV-2 spike in complex with the 1.29 neutraliz...
| Entire | Name: The trimeric SARS-CooV-2 spike in complex with the 1.29 neutralizing nanobody |
|---|---|
| Components |
|
-Supramolecule #1: The trimeric SARS-CooV-2 spike in complex with the 1.29 neutraliz...
| Supramolecule | Name: The trimeric SARS-CooV-2 spike in complex with the 1.29 neutralizing nanobody type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Complex of a soluble spike of the SARS-CoV-2 stabilized in the prefusion form with the 1.29 nanobody bound to its RBD |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.0 kDa/nm |
-Supramolecule #2: SARS-CoV-2 spike
| Supramolecule | Name: SARS-CoV-2 spike / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 / Details: Trimeric spike in its prefusion conformation |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Nanobody 1.29
| Supramolecule | Name: Nanobody 1.29 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 / Details: Immunoglobulin domain |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 140.113922 KDa |
| Recombinant expression | Organism: Mammalian expression vector Flag-MCS-pcDNA3.1 (others) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPQ GFSALEPLVD LPIGINITRF QT LLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCALDPLS ETKCTLKSFT VEKGIYQTSN FRV QPTESI VRFPNITNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNVY ADSF VIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE RDISTEIYQA GSTPC NGVE GFNCYFPLQS YGFQPTNGVG YQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFL PFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ DVNCTEVPVA IHADQLTPTW RVYSTGS NV FQTRAGCLIG AEHVNNSYEC DIPIGAGICA SYQTQTNSPG SASSVASQSI IAYTMSLGAE NSVAYSNNSI AIPTNFTI S VTTEILPVSM TKTSVDCTMY ICGDSTECSN LLLQYGSFCT QLNRALTGIA VEQDKNTQEV FAQVKQIYKT PPIKDFGGF NFSQILPDPS KPSKRSFIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFG AGAALQIPFA MQMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STPSALGKLQ DVVNQNAQAL N TLVKQLSS NFGAISSVLN DILSRLDPPE AEVQIDRLIT GRLQSLQTYV TQQLIRAAEI RASANLAATK MSECVLGQSK RV DFCGKGY HLMSFPQSAP HGVVFLHVTY VPAQEKNFTT APAICHDGKA HFPREGVFVS NGTHWFVTQR NFYEPQIITT DNT FVSGNC DVVIGIVNNT VYDPLQPELD SFKEELDKYF KNHTSPDVDL GDISGINASV VNIQKEIDRL NEVAKNLNES LIDL QELGK YEQGSGSGYI PEAPRDGQAY VRKDGEWVLL STFLGTENLY FQGDYKDDDD KGSHHHHHH UniProtKB: Spike glycoprotein |
-Macromolecule #2: Camel-derived nanobody 1.29
| Macromolecule | Name: Camel-derived nanobody 1.29 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.348559 KDa |
| Recombinant expression | Organism: Mammalian expression vector Flag-MCS-pcDNA3.1 (others) |
| Sequence | String: QVQLVESGGG SVQAGGSLRL SCAASGYTIN TDAVAWFRQA PGKGDERVAV IYTGSGNTNY ADSVKGRFTI SQDNAKNTVY LQMNSLKPE DTALYYCASG YYGASGYDFN NWGQGTQVTV SSALVPR |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 33 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.7 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE | |||||||||
| Details | The spike with the bound nanobody was purified by size exclusion chromatography. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Details | Rigid body refinement. Real space refinement including 5 macro cycles of:Minimization_global, local_grid search and adp. Refinement at 4.0A of resolution. Refinement included NCS. | ||||||||||||
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 140 / Target criteria: correlation coefficient | ||||||||||||
| Output model |  PDB-7r4q: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)