[English] 日本語
 Yorodumi
Yorodumi- EMDB-14032: Structure of the EIAV CA-SP2 hexamer (C2), acquired on Krios G4, ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the EIAV CA-SP2 hexamer (C2), acquired on Krios G4, Selectris/Falcon4, processed with Relion/M | |||||||||
 Map data Map data | EIAV CA hexamer, C2 symmetrized, sharpened and denoised map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Retrovirus / Equine infectious anemia virus / capsid protein / VIRAL PROTEIN | |||||||||
| Biological species |  Equine infectious anemia virus Equine infectious anemia virus | |||||||||
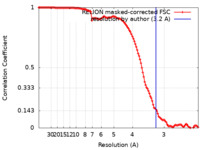
| Method | subtomogram averaging / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Obr M / Hagen WJH / Dick RA / Yu L / Kotecha A / Schur FKM | |||||||||
| Funding support |  Austria, Austria,  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2022 Journal: J Struct Biol / Year: 2022Title: Exploring high-resolution cryo-ET and subtomogram averaging capabilities of contemporary DEDs. Authors: Martin Obr / Wim J H Hagen / Robert A Dick / Lingbo Yu / Abhay Kotecha / Florian K M Schur /     Abstract: The potential of energy filtering and direct electron detection for cryo-electron microscopy (cryo-EM) has been well documented. Here, we assess the performance of recently introduced hardware for ...The potential of energy filtering and direct electron detection for cryo-electron microscopy (cryo-EM) has been well documented. Here, we assess the performance of recently introduced hardware for cryo-electron tomography (cryo-ET) and subtomogram averaging (STA), an increasingly popular structural determination method for complex 3D specimens. We acquired cryo-ET datasets of EIAV virus-like particles (VLPs) on two contemporary cryo-EM systems equipped with different energy filters and direct electron detectors (DED), specifically a Krios G4, equipped with a cold field emission gun (CFEG), Thermo Fisher Scientific Selectris X energy filter, and a Falcon 4 DED; and a Krios G3i, with a Schottky field emission gun (XFEG), a Gatan Bioquantum energy filter, and a K3 DED. We performed constrained cross-correlation-based STA on equally sized datasets acquired on the respective systems. The resulting EIAV CA hexamer reconstructions show that both systems perform comparably in the 4-6 Å resolution range based on Fourier-Shell correlation (FSC). In addition, by employing a recently introduced multiparticle refinement approach, we obtained a reconstruction of the EIAV CA hexamer at 2.9 Å. Our results demonstrate the potential of the new generation of energy filters and DEDs for STA, and the effects of using different processing pipelines on their STA outcomes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14032.map.gz emd_14032.map.gz | 139.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14032-v30.xml emd-14032-v30.xml emd-14032.xml emd-14032.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14032_fsc.xml emd_14032_fsc.xml | 12 KB | Display |  FSC data file FSC data file |
| Images |  emd_14032.png emd_14032.png | 149.5 KB | ||
| Masks |  emd_14032_msk_1.map emd_14032_msk_1.map | 149.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14032.cif.gz emd-14032.cif.gz | 5.8 KB | ||
| Others |  emd_14032_half_map_1.map.gz emd_14032_half_map_1.map.gz emd_14032_half_map_2.map.gz emd_14032_half_map_2.map.gz | 76.9 MB 76.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14032 http://ftp.pdbj.org/pub/emdb/structures/EMD-14032 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14032 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14032 | HTTPS FTP |
-Validation report
| Summary document |  emd_14032_validation.pdf.gz emd_14032_validation.pdf.gz | 636.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14032_full_validation.pdf.gz emd_14032_full_validation.pdf.gz | 636 KB | Display | |
| Data in XML |  emd_14032_validation.xml.gz emd_14032_validation.xml.gz | 18.9 KB | Display | |
| Data in CIF |  emd_14032_validation.cif.gz emd_14032_validation.cif.gz | 24.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14032 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14032 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14032 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14032 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| EM raw data |  EMPIAR-10889 (Title: Tilt series of EIAV CASPNC VLPs acquired on Krios G4 with Selectris X and Falcon 4 EMPIAR-10889 (Title: Tilt series of EIAV CASPNC VLPs acquired on Krios G4 with Selectris X and Falcon 4Data size: 461.4 Data #1: Unaligned movies of EIAV VLPs [micrographs - multiframe] Data #2: Tilt series of EIAV VLPs [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14032.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14032.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EIAV CA hexamer, C2 symmetrized, sharpened and denoised map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.179 Å | ||||||||||||||||||||||||||||||||||||
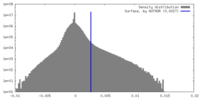
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14032_msk_1.map emd_14032_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: EIAV CA hexamer, C2 symmetrized, half map 1
| File | emd_14032_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EIAV CA hexamer, C2 symmetrized, half map 1 | ||||||||||||
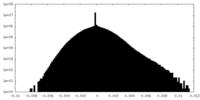
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: EIAV CA hexamer, C2 symmetrized, half map 2
| File | emd_14032_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EIAV CA hexamer, C2 symmetrized, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Equine infectious anemia virus
| Entire | Name:  Equine infectious anemia virus Equine infectious anemia virus |
|---|---|
| Components |
|
-Supramolecule #1: Equine infectious anemia virus
| Supramolecule | Name: Equine infectious anemia virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 11665 / Sci species name: Equine infectious anemia virus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / Name: Immature capsid |
-Macromolecule #1: EIAV CASPNC
| Macromolecule | Name: EIAV CASPNC / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Equine infectious anemia virus Equine infectious anemia virus |
| Recombinant expression | Organism:  |
| Sequence | String: SPIMIDGAGN RNFRPLTPRG YTTWVNTIQT NGLLNEASQN LFGILSVDCT SEEMNAFLDV V PGQAGQKQ ILLDAIDKIA DDWDNRHPLP NAPLVAPPQG PIPMTARFIR GLGVPRERQM EP AFDQFRQ TYRQWIIEAM SEGIKVMIGK PKAQNIRQGA KEPYPEFVDR ...String: SPIMIDGAGN RNFRPLTPRG YTTWVNTIQT NGLLNEASQN LFGILSVDCT SEEMNAFLDV V PGQAGQKQ ILLDAIDKIA DDWDNRHPLP NAPLVAPPQG PIPMTARFIR GLGVPRERQM EP AFDQFRQ TYRQWIIEAM SEGIKVMIGK PKAQNIRQGA KEPYPEFVDR LLSQIKSEGH PQE ISKFLT DTLTIQNANE ECRNAMRHLR PEDTLEEKMY ACRDIGTTKQ KMML LAKAL QTGLAGPFK GGALKGGPLK AAQTCYNCGK PGHLSSQCRA PKVCFKCKQP GHFSKQCRSV P KNGKQGAQ GRPQKQTF |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.2 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Instrument: LEICA EM GP Details: 3.5 seconds blotting time, using back-side blotting. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Average exposure time: 1.02 sec. / Average electron dose: 3.5 e/Å2 / Details: Images acquired in the EER format. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.25 µm / Nominal defocus min: 0.75 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)