[English] 日本語
 Yorodumi
Yorodumi- EMDB-13968: Cryo-EM structure of the Tripartite ATP-independent Periplasmic (... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in amphipol | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Photobacterium profundum (bacteria) / Photobacterium profundum (bacteria) /   Photobacterium profundum SS9 (bacteria) / synthetic construct (others) Photobacterium profundum SS9 (bacteria) / synthetic construct (others) | |||||||||
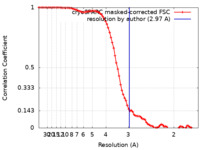
| Method | single particle reconstruction / cryo EM / Resolution: 2.97 Å | |||||||||
 Authors Authors | North RA / Davies JS / Morado D / Dobson RCJ | |||||||||
| Funding support |  New Zealand, 2 items New Zealand, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter. Authors: James S Davies / Michael J Currie / Rachel A North / Mariafrancesca Scalise / Joshua D Wright / Jack M Copping / Daniela M Remus / Ashutosh Gulati / Dustin R Morado / Sam A Jamieson / ...Authors: James S Davies / Michael J Currie / Rachel A North / Mariafrancesca Scalise / Joshua D Wright / Jack M Copping / Daniela M Remus / Ashutosh Gulati / Dustin R Morado / Sam A Jamieson / Michael C Newton-Vesty / Gayan S Abeysekera / Subramanian Ramaswamy / Rosmarie Friemann / Soichi Wakatsuki / Jane R Allison / Cesare Indiveri / David Drew / Peter D Mace / Renwick C J Dobson /      Abstract: In bacteria and archaea, tripartite ATP-independent periplasmic (TRAP) transporters uptake essential nutrients. TRAP transporters receive their substrates via a secreted soluble substrate-binding ...In bacteria and archaea, tripartite ATP-independent periplasmic (TRAP) transporters uptake essential nutrients. TRAP transporters receive their substrates via a secreted soluble substrate-binding protein. How a sodium ion-driven secondary active transporter is strictly coupled to a substrate-binding protein is poorly understood. Here we report the cryo-EM structure of the sialic acid TRAP transporter SiaQM from Photobacterium profundum at 2.97 Å resolution. SiaM comprises a "transport" domain and a "scaffold" domain, with the transport domain consisting of helical hairpins as seen in the sodium ion-coupled elevator transporter VcINDY. The SiaQ protein forms intimate contacts with SiaM to extend the size of the scaffold domain, suggesting that TRAP transporters may operate as monomers, rather than the typically observed oligomers for elevator-type transporters. We identify the Na and sialic acid binding sites in SiaM and demonstrate a strict dependence on the substrate-binding protein SiaP for uptake. We report the SiaP crystal structure that, together with docking studies, suggest the molecular basis for how sialic acid is delivered to the SiaQM transporter complex. We thus propose a model for substrate transport by TRAP proteins, which we describe herein as an 'elevator-with-an-operator' mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13968.map.gz emd_13968.map.gz | 26.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13968-v30.xml emd-13968-v30.xml emd-13968.xml emd-13968.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13968_fsc.xml emd_13968_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_13968.png emd_13968.png | 69.8 KB | ||
| Masks |  emd_13968_msk_1.map emd_13968_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Others |  emd_13968_half_map_1.map.gz emd_13968_half_map_1.map.gz emd_13968_half_map_2.map.gz emd_13968_half_map_2.map.gz | 48.9 MB 48.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13968 http://ftp.pdbj.org/pub/emdb/structures/EMD-13968 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13968 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13968 | HTTPS FTP |
-Validation report
| Summary document |  emd_13968_validation.pdf.gz emd_13968_validation.pdf.gz | 749.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13968_full_validation.pdf.gz emd_13968_full_validation.pdf.gz | 749.2 KB | Display | |
| Data in XML |  emd_13968_validation.xml.gz emd_13968_validation.xml.gz | 15.3 KB | Display | |
| Data in CIF |  emd_13968_validation.cif.gz emd_13968_validation.cif.gz | 19 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13968 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13968 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13968 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13968 | HTTPS FTP |
-Related structure data
| Related structure data |  7qhaMC  7t3eC  8b01C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13968.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13968.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83063 Å | ||||||||||||||||||||||||||||||||||||
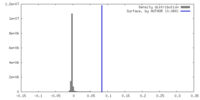
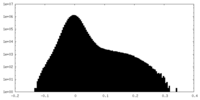
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13968_msk_1.map emd_13968_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
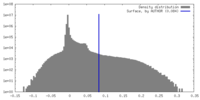
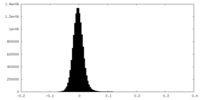
| Density Histograms |
-Half map: #2
| File | emd_13968_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
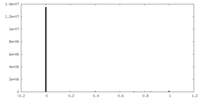
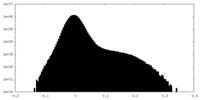
| Density Histograms |
-Half map: #1
| File | emd_13968_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
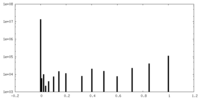
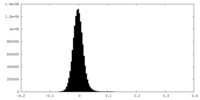
| Density Histograms |
- Sample components
Sample components
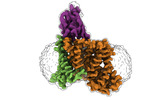
-Entire : SiaQM with megabody
| Entire | Name: SiaQM with megabody |
|---|---|
| Components |
|
-Supramolecule #1: SiaQM with megabody
| Supramolecule | Name: SiaQM with megabody / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: SiaQM
| Supramolecule | Name: SiaQM / type: complex / ID: 2 / Chimera: Yes / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Photobacterium profundum (bacteria) Photobacterium profundum (bacteria) |
-Supramolecule #3: Megabody
| Supramolecule | Name: Megabody / type: complex / ID: 3 / Chimera: Yes / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Putative TRAP-type C4-dicarboxylate transport system, small perme...
| Macromolecule | Name: Putative TRAP-type C4-dicarboxylate transport system, small permease component type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Photobacterium profundum SS9 (bacteria) / Strain: SS9 Photobacterium profundum SS9 (bacteria) / Strain: SS9 |
| Molecular weight | Theoretical: 19.748367 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFRKIIDNIE EIITVPLMIA LLCILTWQIS SRWLFDSPSL WSEELARVLF LHMAIIGGAI AIKKDDHVKI TFFSDKLPRN FRYSLLFAL ELLVLITIVA MIYYGYAHVQ RTAFFELITL GISSSWMTYA LPVGGCFMLV RQCQKLYFVL IDWRINENKN T SHLTACDI NE |
-Macromolecule #2: Putative TRAP-type C4-dicarboxylate transport system, large perme...
| Macromolecule | Name: Putative TRAP-type C4-dicarboxylate transport system, large permease component type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Photobacterium profundum SS9 (bacteria) / Strain: SS9 Photobacterium profundum SS9 (bacteria) / Strain: SS9 |
| Molecular weight | Theoretical: 45.573605 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFTSIVGWLG LLFAGMPVGF SLIFVGLAFL VLTESTGINF AAQQMIGGLD NFTLLAVPFF VLTGHLMNSA GITERIFNFA KAMVGHITG SLGHVNILAS LLFSGMSGSA LADAGGLGQL EIKSMRDAKY DDDFAGGLTA ASCIIGPLVP PSIPLVIYGV V SNTSIGAL ...String: MFTSIVGWLG LLFAGMPVGF SLIFVGLAFL VLTESTGINF AAQQMIGGLD NFTLLAVPFF VLTGHLMNSA GITERIFNFA KAMVGHITG SLGHVNILAS LLFSGMSGSA LADAGGLGQL EIKSMRDAKY DDDFAGGLTA ASCIIGPLVP PSIPLVIYGV V SNTSIGAL FLAGAIPGLL CCIALCIMTY FIAKKRGYMT LPRASRKERL IAFRDAFLSL LTPFIIIGGI FSGKFTPTEA AI ISSLYAL FLGTVVYKSL TMDKFIKLVQ ETVTTTSVVA LMVMGVTVFG WIVAREQLPQ QLAELFLSIS DNPLILLLLI NLL LLFLGT FIESLALLLL LVPFLVPVAT SVGIDPVHFG VMAILNLMIG ILTPPMGMAL YVVSKVGNIP FHVLTRGVLP LLVP LFIVL GLIIVFPQIT LFLPQLVLGY GL |
-Macromolecule #3: Megabody c7HopQ
| Macromolecule | Name: Megabody c7HopQ / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 54.867098 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQTKTTTSV IDTTNDAQNL LTQAQTIVNT LKDYCPILIA KSSSSNGGTN NANTPSWQTA GGGKNSCATF GAEFSAASD MINNAQKIVQ ETQQLSANQP KNITQPHNLN LNSPSSLTAL AQKMLKNAQS QAEILKLANQ VESDFNKLSS G HLKDYIGK ...String: QVQLQESGGG LVQTKTTTSV IDTTNDAQNL LTQAQTIVNT LKDYCPILIA KSSSSNGGTN NANTPSWQTA GGGKNSCATF GAEFSAASD MINNAQKIVQ ETQQLSANQP KNITQPHNLN LNSPSSLTAL AQKMLKNAQS QAEILKLANQ VESDFNKLSS G HLKDYIGK CDASAISSAN MTMQNQKNNW GNGCAGVEET QSLLKTSAAD FNNQTPQINQ AQNLANTLIQ ELGNNDTYEQ LS RLLTNDN GTNSKTSAQA INQAVNNLNE RAKTLAGGTT NSPAYQATLL ALRSVLGLWN SMGYAVICGG YTKSPGENNQ KDF HYTDEN GNGTTINCGG STNSNGTHSY NGTNTLKADK NVSLSIEQYE KIHEAYQILS KALKQAGLAP LNSKGEKLEA HVTT SKYAG GSLRLSCAAS GNIFDRGYMG WYRQAPGKER ELVAGISYGG STYYADSVKG RFTISRDNAK NTVYLQMNSL KPEDT AVYY CAAYPLYDDP YYYWGQGTQV TVSSLE |
-Macromolecule #4: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 4 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.9 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 10960 / Average exposure time: 1.9 sec. / Average electron dose: 71.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.4 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)