+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of full-length human immunoglobulin M | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
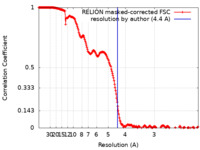
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||||||||||||||
 Authors Authors | Chen Q / Rosenthal P / Tolar P | |||||||||||||||||||||
| Funding support |  United Kingdom, 6 items United Kingdom, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer. Authors: Qu Chen / Rajesh Menon / Lesley J Calder / Pavel Tolar / Peter B Rosenthal /  Abstract: Immunoglobulin M (IgM) is the most ancient of the five isotypes of immunoglobulin (Ig) molecules and serves as the first line of defence against pathogens. Here, we use cryo-EM to image the structure ...Immunoglobulin M (IgM) is the most ancient of the five isotypes of immunoglobulin (Ig) molecules and serves as the first line of defence against pathogens. Here, we use cryo-EM to image the structure of the human full-length IgM pentamer, revealing antigen binding domains flexibly attached to the asymmetric and rigid core formed by the Cμ4 and Cμ3 constant regions and the J-chain. A hinge is located at the Cμ3/Cμ2 domain interface, allowing Fabs and Cμ2 to pivot as a unit both in-plane and out-of-plane. This motion is different from that observed in IgG and IgA, where the two Fab arms are able to swing independently. A biased orientation of one pair of Fab arms results from asymmetry in the constant domain (Cμ3) at the IgM subunit interacting most extensively with the J-chain. This may influence the multi-valent binding to surface-associated antigens and complement pathway activation. By comparison, the structure of the Fc fragment in the IgM monomer is similar to that of the pentamer, but is more dynamic in the Cμ4 domain. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13921.map.gz emd_13921.map.gz | 778.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13921-v30.xml emd-13921-v30.xml emd-13921.xml emd-13921.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13921_fsc.xml emd_13921_fsc.xml | 21.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_13921.png emd_13921.png | 134.7 KB | ||
| Masks |  emd_13921_msk_1.map emd_13921_msk_1.map | 824 MB |  Mask map Mask map | |
| Others |  emd_13921_additional_1.map.gz emd_13921_additional_1.map.gz emd_13921_half_map_1.map.gz emd_13921_half_map_1.map.gz emd_13921_half_map_2.map.gz emd_13921_half_map_2.map.gz | 410.5 MB 764.8 MB 764.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13921 http://ftp.pdbj.org/pub/emdb/structures/EMD-13921 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13921 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13921 | HTTPS FTP |
-Validation report
| Summary document |  emd_13921_validation.pdf.gz emd_13921_validation.pdf.gz | 638.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13921_full_validation.pdf.gz emd_13921_full_validation.pdf.gz | 637.9 KB | Display | |
| Data in XML |  emd_13921_validation.xml.gz emd_13921_validation.xml.gz | 28.9 KB | Display | |
| Data in CIF |  emd_13921_validation.cif.gz emd_13921_validation.cif.gz | 38.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13921 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13921 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13921 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13921 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13921.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13921.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13921_msk_1.map emd_13921_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_13921_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_13921_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_13921_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Full-length human IgM pentamer
| Entire | Name: Full-length human IgM pentamer |
|---|---|
| Components |
|
-Supramolecule #1: Full-length human IgM pentamer
| Supramolecule | Name: Full-length human IgM pentamer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: J chain
| Macromolecule | Name: J chain / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QEDERIVLVD NKCKCARITS RIIRSSEDPN EDIVERNIRI IVPLNNRENI SDPTSPLRTR FVYHLSDLCK KCDPTEVELD NQIVTATQSN ICDEDSATET CYTYDRNKCY TAVVPLVYGG ETKMVETALT PDACY |
-Macromolecule #2: human IgM heavy chain
| Macromolecule | Name: human IgM heavy chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVTLKESGPT LVKPTQTLTL TCTFSGFSLT TTGEGVGWIR QPPGKALEFL AFIYWNDAKR YNPSLQSRLT ITKDASKKQV VLTLTNLDPV DTATYYCART SGWDIEFEYW GQGTLVTVSS GSASAPTLFP LVSCENSPSD TSSVAVGCLA QDFLPDSITF SWKYKNNSDI ...String: QVTLKESGPT LVKPTQTLTL TCTFSGFSLT TTGEGVGWIR QPPGKALEFL AFIYWNDAKR YNPSLQSRLT ITKDASKKQV VLTLTNLDPV DTATYYCART SGWDIEFEYW GQGTLVTVSS GSASAPTLFP LVSCENSPSD TSSVAVGCLA QDFLPDSITF SWKYKNNSDI SSTRGFPSVL RGGKYAATSQ VLLPSKDVMQ GTDEHVVCKV QHPNGNKEKN VPLPVIAELP PKVSVFVPPR DGFFGNPRKS KLICQATGFS PRQIQVSWLR EGKQVGSGVT TDQVQAEAKE SGPTTYKVTS TLTIKESDWL GQSMFTCRVD HRGLTFQQNA SSMCVPDQDT AIRVFAIPPS FASIFLTKST KLTCLVTDLT TYDSVTISWT RQNGEAVKTH TNISESHPNA TFSAVGEASI CEDDWNSGER FTCTVTHTDL PSPLKQTISR PKGVALHRPD VYLLPPAREQ LNLRESATIT CLVTGFSPAD VFVQWMQRGQ PLSPEKYVTS AMPEPQAPGR YFAHSILTVS EEEWNTGETY TCVVAHEALP NRVTERTVDK STGKPTLYNV SLVMSDTAGT CY |
-Macromolecule #3: human IgM light chain
| Macromolecule | Name: human IgM light chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGERAT LSCRASETVS NDKVAWYQQK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLSISGLE PEDFVVYYCQ QYASSPRTFG QGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS QESVTEQDSK ...String: EIVLTQSPGT LSLSPGERAT LSCRASETVS NDKVAWYQQK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLSISGLE PEDFVVYYCQ QYASSPRTFG QGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS QESVTEQDSK DSTYSLSSTL TLSKADYEKH KVYACEVTHQ GLSSPVTKSF NRGEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 34.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 5.0 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 128440 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)