[English] 日本語
 Yorodumi
Yorodumi- EMDB-13876: Cryo-EM structure of the octameric pore of Clostridium perfringen... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
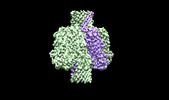
| Title | Cryo-EM structure of the octameric pore of Clostridium perfringens beta-toxin. | |||||||||
 Map data Map data | postprocessed map CPB | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | pore forming toxin / hemolysin / octamer / TOXIN | |||||||||
| Biological species |   Clostridium perfringens CPE (bacteria) Clostridium perfringens CPE (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.84 Å | |||||||||
 Authors Authors | Iacovache I / Zuber B | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2022 Journal: EMBO Rep / Year: 2022Title: Cryo-EM structure of the octameric pore of Clostridium perfringens β-toxin. Authors: Julia Bruggisser / Ioan Iacovache / Samuel C Musson / Matteo T Degiacomi / Horst Posthaus / Benoît Zuber /   Abstract: Clostridium perfringens is one of the most widely distributed and successful pathogens producing an impressive arsenal of toxins. One of the most potent toxins produced is the C. perfringens β-toxin ...Clostridium perfringens is one of the most widely distributed and successful pathogens producing an impressive arsenal of toxins. One of the most potent toxins produced is the C. perfringens β-toxin (CPB). This toxin is the main virulence factor of type C strains. We describe the cryo-electron microscopy (EM) structure of CPB oligomer. We show that CPB forms homo-octameric pores like the hetero-oligomeric pores of the bi-component leukocidins, with important differences in the receptor binding region and the N-terminal latch domain. Intriguingly, the octameric CPB pore complex contains a second 16-stranded β-barrel protrusion atop of the cap domain that is formed by the N-termini of the eight protomers. We propose that CPB, together with the newly identified Epx toxins, is a member a new subclass of the hemolysin-like family. In addition, we show that the β-barrel protrusion domain can be modified without affecting the pore-forming ability, thus making the pore particularly attractive for macromolecule sensing and nanotechnology. The cryo-EM structure of the octameric pore of CPB will facilitate future developments in both nanotechnology and basic research. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13876.map.gz emd_13876.map.gz | 4.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13876-v30.xml emd-13876-v30.xml emd-13876.xml emd-13876.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13876_fsc.xml emd_13876_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_13876.png emd_13876.png | 47.4 KB | ||
| Filedesc metadata |  emd-13876.cif.gz emd-13876.cif.gz | 5.6 KB | ||
| Others |  emd_13876_half_map_1.map.gz emd_13876_half_map_1.map.gz emd_13876_half_map_2.map.gz emd_13876_half_map_2.map.gz | 20.1 MB 20.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13876 http://ftp.pdbj.org/pub/emdb/structures/EMD-13876 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13876 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13876 | HTTPS FTP |
-Validation report
| Summary document |  emd_13876_validation.pdf.gz emd_13876_validation.pdf.gz | 649.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13876_full_validation.pdf.gz emd_13876_full_validation.pdf.gz | 649.3 KB | Display | |
| Data in XML |  emd_13876_validation.xml.gz emd_13876_validation.xml.gz | 13 KB | Display | |
| Data in CIF |  emd_13876_validation.cif.gz emd_13876_validation.cif.gz | 18 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13876 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13876 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13876 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13876 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13876.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13876.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocessed map CPB | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.306 Å | ||||||||||||||||||||||||||||||||||||
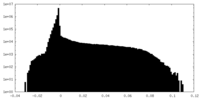
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half1
| File | emd_13876_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
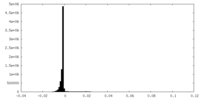
| Density Histograms |
-Half map: half2
| File | emd_13876_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : octamer of beta toxin in SMALP
| Entire | Name: octamer of beta toxin in SMALP |
|---|---|
| Components |
|
-Supramolecule #1: octamer of beta toxin in SMALP
| Supramolecule | Name: octamer of beta toxin in SMALP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 250 KDa |
-Macromolecule #1: Clostridium perfringens beta toxin
| Macromolecule | Name: Clostridium perfringens beta toxin / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Clostridium perfringens CPE (bacteria) Clostridium perfringens CPE (bacteria) |
| Molecular weight | Theoretical: 34.89375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NDIGKTTTIT RNKTSDGYTI ITQNDKQIIS YQSVDSSSKN EDGFTASIDA RFIDDKYSSE MTTLINLTGF MSSKKEDVIK KYNLHDVTN STAINFPVRY SISILNESIN ENVKIVDSIP KNTISQKTVS NTMGYKIGGS IEIEENKPKA SIESEYAESS T IEYVQPDF ...String: NDIGKTTTIT RNKTSDGYTI ITQNDKQIIS YQSVDSSSKN EDGFTASIDA RFIDDKYSSE MTTLINLTGF MSSKKEDVIK KYNLHDVTN STAINFPVRY SISILNESIN ENVKIVDSIP KNTISQKTVS NTMGYKIGGS IEIEENKPKA SIESEYAESS T IEYVQPDF STIQTDHSTS KASWDTKFTE TTRGNYNLKS NNPVYGNEMF MYGRYTNVPA TENIIPDYQM SKLITGGLNP NM SVVLTAP NGTEESIIKV KMERERNCYY LNWNGANWVG QVYSRLAFDT PNVDSHIFTF KINWLTHKVT AI |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Component:
| ||||||
|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.25 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.8 µm |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)