[English] 日本語
 Yorodumi
Yorodumi- EMDB-1283: Molecular architecture and conformational flexibility of human RN... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1283 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Molecular architecture and conformational flexibility of human RNA polymerase II. | |||||||||
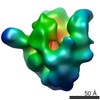
 Map data Map data | Conformation 1 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 20.0 Å | |||||||||
 Authors Authors | Kostek SA / Grob P / De Carlo S / Lipscomb JS / Garczarek F / Nogales E | |||||||||
 Citation Citation |  Journal: Structure / Year: 2006 Journal: Structure / Year: 2006Title: Molecular architecture and conformational flexibility of human RNA polymerase II. Authors: Seth A Kostek / Patricia Grob / Sacha De Carlo / J Slaton Lipscomb / Florian Garczarek / Eva Nogales /  Abstract: Transcription by RNA polymerase II (RNAPII) is a central process in eukaryotic gene regulation. While atomic details exist for the yeast RNAPII, characterization of the human complex lags behind, ...Transcription by RNA polymerase II (RNAPII) is a central process in eukaryotic gene regulation. While atomic details exist for the yeast RNAPII, characterization of the human complex lags behind, mostly due to the inability to obtain large quantities of purified material. Although the complexes have the same protein composition and high sequence similarity, understanding of transcription and of transcription-coupled DNA repair (TCR) in humans will require the use of human proteins in structural studies. We have used cryo-electron microscopy, image reconstruction, and variance analysis to characterize the structure and dynamics of human RNAPII (hRNAPII). Our studies show that hRNAPII in solution parallels the conformational flexibility of the yeast structures crystallized in different states but also illustrate a more extensive conformational range with potential biological significance. This hRNAPII study will serve as a structural platform to build up higher-order transcription and TCR complexes and to gain information that may be unique to the human RNAPII system. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1283.map.gz emd_1283.map.gz | 5.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1283-v30.xml emd-1283-v30.xml emd-1283.xml emd-1283.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| Images |  1283.gif 1283.gif | 21.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1283 http://ftp.pdbj.org/pub/emdb/structures/EMD-1283 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1283 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1283 | HTTPS FTP |
-Validation report
| Summary document |  emd_1283_validation.pdf.gz emd_1283_validation.pdf.gz | 202 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1283_full_validation.pdf.gz emd_1283_full_validation.pdf.gz | 201.1 KB | Display | |
| Data in XML |  emd_1283_validation.xml.gz emd_1283_validation.xml.gz | 4.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1283 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1283 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1283 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1283 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1283.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1283.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Conformation 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : human RNA Polymerase II
+Supramolecule #1000: human RNA Polymerase II
+Macromolecule #1: Rpb1
+Macromolecule #2: Rpb2
+Macromolecule #3: Rpb3
+Macromolecule #4: Rpb4
+Macromolecule #5: Rpb5
+Macromolecule #6: Rpb6
+Macromolecule #7: Rpb7
+Macromolecule #8: Rpb8
+Macromolecule #9: Rpb9
+Macromolecule #10: Rpb10
+Macromolecule #11: Rpb11
+Macromolecule #12: Rpb12
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.9 Details: 200mM (NH4)2SO4, 25mM Hepes, 0.2M EDTA, 0.05% NP-40 |
| Staining | Type: NEGATIVE Details: cryo-negative staining in a saturated solution of ammonium molybdate neutralized to a pH of 7.2. with 10 N NaOH |
| Grid | Details: 400 mesh copper grid |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 90 K / Method: stained with ammonium molybdate before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Temperature | Average: 90 K |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 6.3 µm / Number real images: 19 / Average electron dose: 17 e/Å2 / Od range: 1 / Bits/pixel: 14 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 50280 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus max: 3.6 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: side entry / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: each micrograph |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: FREALIGN / Number images used: 3406 |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















