[English] 日本語
 Yorodumi
Yorodumi- EMDB-12539: Tomogram of mutant huntingtin containing organelle resembles auto... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12539 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
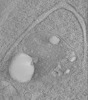
| Title | Tomogram of mutant huntingtin containing organelle resembles autolysosome/residual body in 6-month-old zQ175 mice | |||||||||||||||
 Map data Map data | Tomogram of mutant HTT containing organelle resembles autolysosome/residual body in 6-month-old zQ175 mice. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | electron tomography / negative staining | |||||||||||||||
 Authors Authors | Zhou Y / Saibil HR | |||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Acta Neuropathol Commun / Year: 2021 Journal: Acta Neuropathol Commun / Year: 2021Title: Correlative light and electron microscopy suggests that mutant huntingtin dysregulates the endolysosomal pathway in presymptomatic Huntington's disease. Authors: Ya Zhou / Thomas R Peskett / Christian Landles / John B Warner / Kirupa Sathasivam / Edward J Smith / Shu Chen / Ronald Wetzel / Hilal A Lashuel / Gillian P Bates / Helen R Saibil /    Abstract: Huntington's disease (HD) is a late onset, inherited neurodegenerative disorder for which early pathogenic events remain poorly understood. Here we show that mutant exon 1 HTT proteins are recruited ...Huntington's disease (HD) is a late onset, inherited neurodegenerative disorder for which early pathogenic events remain poorly understood. Here we show that mutant exon 1 HTT proteins are recruited to a subset of cytoplasmic aggregates in the cell bodies of neurons in brain sections from presymptomatic HD, but not wild-type, mice. This occurred in a disease stage and polyglutamine-length dependent manner. We successfully adapted a high-resolution correlative light and electron microscopy methodology, originally developed for mammalian and yeast cells, to allow us to correlate light microscopy and electron microscopy images on the same brain section within an accuracy of 100 nm. Using this approach, we identified these recruitment sites as single membrane bound, vesicle-rich endolysosomal organelles, specifically as (1) multivesicular bodies (MVBs), or amphisomes and (2) autolysosomes or residual bodies. The organelles were often found in close-proximity to phagophore-like structures. Immunogold labeling localized mutant HTT to non-fibrillar, electron lucent structures within the lumen of these organelles. In presymptomatic HD, the recruitment organelles were predominantly MVBs/amphisomes, whereas in late-stage HD, there were more autolysosomes or residual bodies. Electron tomograms indicated the fusion of small vesicles with the vacuole within the lumen, suggesting that MVBs develop into residual bodies. We found that markers of MVB-related exocytosis were depleted in presymptomatic mice and throughout the disease course. This suggests that endolysosomal homeostasis has moved away from exocytosis toward lysosome fusion and degradation, in response to the need to clear the chronically aggregating mutant HTT protein, and that this occurs at an early stage in HD pathogenesis. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12539.map.gz emd_12539.map.gz | 2.4 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12539-v30.xml emd-12539-v30.xml emd-12539.xml emd-12539.xml | 10.8 KB 10.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_12539.png emd_12539.png | 126.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12539 http://ftp.pdbj.org/pub/emdb/structures/EMD-12539 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12539 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12539 | HTTPS FTP |
-Validation report
| Summary document |  emd_12539_validation.pdf.gz emd_12539_validation.pdf.gz | 181.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12539_full_validation.pdf.gz emd_12539_full_validation.pdf.gz | 181 KB | Display | |
| Data in XML |  emd_12539_validation.xml.gz emd_12539_validation.xml.gz | 4.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12539 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12539 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12539 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12539 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12539.map.gz / Format: CCP4 / Size: 4.7 GB / Type: IMAGE STORED AS SIGNED BYTE Download / File: emd_12539.map.gz / Format: CCP4 / Size: 4.7 GB / Type: IMAGE STORED AS SIGNED BYTE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tomogram of mutant HTT containing organelle resembles autolysosome/residual body in 6-month-old zQ175 mice. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.441 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Mutant huntingtin containing organelle in hippocampal CA1 neuron ...
| Entire | Name: Mutant huntingtin containing organelle in hippocampal CA1 neuron from zQ175 knock-in mice. |
|---|---|
| Components |
|
-Supramolecule #1: Mutant huntingtin containing organelle in hippocampal CA1 neuron ...
| Supramolecule | Name: Mutant huntingtin containing organelle in hippocampal CA1 neuron from zQ175 knock-in mice. type: tissue / ID: 1 / Parent: 0 Details: Cytoplasmic inclusion sites identified by correlative fluorescence/EM on freeze substituted brain sections. |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | tissue |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Details: MilliQ water was used. |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl acetate |
| Sugar embedding | Material: Lowicryl HM20 Details: High pressure frozen samples were freeze substituted in a Leica AFS |
| Details | Brain slices were high pressure frozen and freeze substituted |
| High pressure freezing | Instrument: OTHER Details: The value given for _emd_high_pressure_freezing.instrument is HPM100. This is not in a list of allowed values {'EMS-002 RAPID IMMERSION FREEZER', 'OTHER', 'LEICA EM PACT', 'LEICA EM HPM100', ...Details: The value given for _emd_high_pressure_freezing.instrument is HPM100. This is not in a list of allowed values {'EMS-002 RAPID IMMERSION FREEZER', 'OTHER', 'LEICA EM PACT', 'LEICA EM HPM100', 'LEICA EM PACT2', 'BAL-TEC HPM 010'} so OTHER is written into the XML file. |
| Cryo protectant | BSA |
| Sectioning | Ultramicrotomy - Instrument: Leica EM UC7 / Ultramicrotomy - Temperature: 20 K / Ultramicrotomy - Final thickness: 300 nm |
| Fiducial marker | Manufacturer: EMS / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 19.38 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Software - Name:  IMOD / Number images used: 37 IMOD / Number images used: 37 |
|---|
 Movie
Movie Controller
Controller