[English] 日本語
 Yorodumi
Yorodumi- EMDB-11156: Plasmodium falciparum merozoite surface protein 1 dimer, conforma... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11156 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Plasmodium falciparum merozoite surface protein 1 dimer, conformation 1 | |||||||||
 Map data Map data | Dimer, C2 reconstruction, unfiltered | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationvacuolar membrane / membrane => GO:0016020 / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
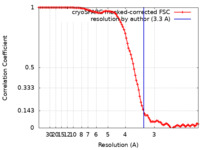
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Dijkman PM / Kudryashev M | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Structure of the merozoite surface protein 1 from . Authors: Patricia M Dijkman / Tanja Marzluf / Yingyi Zhang / Shih-Ying Scott Chang / Dominic Helm / Michael Lanzer / Hermann Bujard / Mikhail Kudryashev /  Abstract: The merozoite surface protein 1 (MSP-1) is the most abundant protein on the surface of the erythrocyte-invading merozoite, the causative agent of malaria. MSP-1 is essential for merozoite formation, ...The merozoite surface protein 1 (MSP-1) is the most abundant protein on the surface of the erythrocyte-invading merozoite, the causative agent of malaria. MSP-1 is essential for merozoite formation, entry into and escape from erythrocytes, and is a promising vaccine candidate. Here, we present monomeric and dimeric structures of full-length MSP-1. MSP-1 adopts an unusual fold with a large central cavity. Its fold includes several coiled-coils and shows structural homology to proteins associated with membrane and cytoskeleton interactions. MSP-1 formed dimers through these domains in a concentration-dependent manner. Dimerization is affected by the presence of the erythrocyte cytoskeleton protein spectrin, which may compete for the dimerization interface. Our work provides structural insights into the possible mode of interaction of MSP-1 with erythrocytes and establishes a framework for future investigations into the role of MSP-1 in infection and immunity. #1:  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Structure of the merozoite surface protein 1 from Plasmodium falciparum Authors: Dijkman PM / Marzluf T / Zhang Y / Chang SYS / Helm D / Lanzer M / Bujard H / Kudryashev M | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11156.map.gz emd_11156.map.gz | 45.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11156-v30.xml emd-11156-v30.xml emd-11156.xml emd-11156.xml | 31 KB 31 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11156_fsc.xml emd_11156_fsc.xml | 10.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_11156.png emd_11156.png | 132.3 KB | ||
| Masks |  emd_11156_msk_1.map emd_11156_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Others |  emd_11156_additional_1.map.gz emd_11156_additional_1.map.gz emd_11156_additional_2.map.gz emd_11156_additional_2.map.gz emd_11156_additional_3.map.gz emd_11156_additional_3.map.gz emd_11156_additional_4.map.gz emd_11156_additional_4.map.gz emd_11156_additional_5.map.gz emd_11156_additional_5.map.gz emd_11156_half_map_1.map.gz emd_11156_half_map_1.map.gz emd_11156_half_map_2.map.gz emd_11156_half_map_2.map.gz | 86 MB 71 MB 6.8 MB 71.1 MB 71.3 MB 84.5 MB 84.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11156 http://ftp.pdbj.org/pub/emdb/structures/EMD-11156 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11156 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11156 | HTTPS FTP |
-Validation report
| Summary document |  emd_11156_validation.pdf.gz emd_11156_validation.pdf.gz | 466.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11156_full_validation.pdf.gz emd_11156_full_validation.pdf.gz | 465.7 KB | Display | |
| Data in XML |  emd_11156_validation.xml.gz emd_11156_validation.xml.gz | 17.8 KB | Display | |
| Data in CIF |  emd_11156_validation.cif.gz emd_11156_validation.cif.gz | 22.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11156 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11156 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11156 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11156 | HTTPS FTP |
-Related structure data
| Related structure data |  6zbjMC  6zbcC  6zbdC  6zbeC  6zbfC  6zbgC  6zbhC  6zblC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10437 (Title: Single-particle cryo-EM of the full-length merozoite surface protein 1 from Plasmodium falciparum EMPIAR-10437 (Title: Single-particle cryo-EM of the full-length merozoite surface protein 1 from Plasmodium falciparumData size: 34.0 TB Data #1: Unaligned movies of MSP-1 [micrographs - multiframe] Data #2: Unaligned movies of hdMSP-1 (dataset 1) [micrographs - multiframe] Data #3: Unaligned movies of hdMSP-1 (dataset 2) [micrographs - multiframe] Data #4: Unaligned movies of hdMSP-1 (dataset 3) [micrographs - multiframe] Data #5: Unaligned movies of hdMSP-1 (dataset 4) [micrographs - multiframe] Data #6: Final particle stack for hdMSP-1 conformation 1 [picked particles - single frame - processed] Data #7: Final particle stack for hdMSP-1 conformation 2 [picked particles - single frame - processed] Data #8: Final particle stack for MSP-1 main conformation [picked particles - single frame - processed] Data #9: Final particle stack for MSP-1 alternative conformation 1 [picked particles - single frame - processed] Data #10: Final particle stack for MSP-1 alternative conformation 2 [picked particles - single frame - processed] Data #11: Final particle stack for MSP-1 alternative conformation 3 [picked particles - single frame - processed] Data #12: Final particle stack for MSP-1 alternative conformation 4 [picked particles - single frame - processed] Data #13: Final particle stack for MSP-1 alternative conformation 5 [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11156.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11156.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Dimer, C2 reconstruction, unfiltered | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.077 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
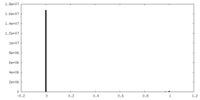
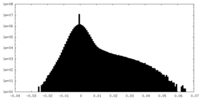
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11156_msk_1.map emd_11156_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
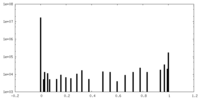
| Projections & Slices |
| ||||||||||||
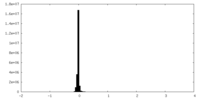
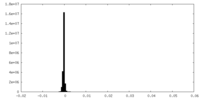
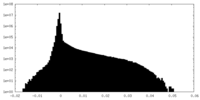
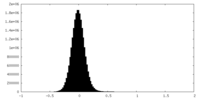
| Density Histograms |
-Additional map: Dimer, C2 reconstruction, sharpened
| File | emd_11156_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Dimer, C2 reconstruction, sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
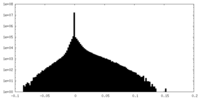
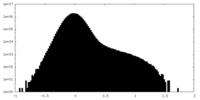
| Density Histograms |
-Additional map: C2 symmetry expansion, C1 monomer reconstruction, unfiltered
| File | emd_11156_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C2 symmetry expansion, C1 monomer reconstruction, unfiltered | ||||||||||||
| Projections & Slices |
| ||||||||||||
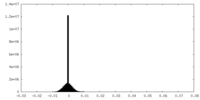
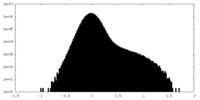
| Density Histograms |
-Additional map: C2 symmetry expansion, C1 monomer reconstruction, globally sharpened
| File | emd_11156_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C2 symmetry expansion, C1 monomer reconstruction, globally sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
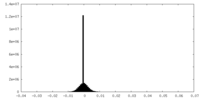
| Density Histograms |
-Additional map: C2 symmetry expansion, C1 monomer reconstruction, half map 2
| File | emd_11156_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C2 symmetry expansion, C1 monomer reconstruction, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: C2 symmetry expansion, C1 monomer reconstruction, half map 1
| File | emd_11156_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C2 symmetry expansion, C1 monomer reconstruction, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: C2 dimer, half map 1
| File | emd_11156_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C2 dimer, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: C2 dimer, half map 2
| File | emd_11156_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C2 dimer, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Merozoite surface protein 1 (MSP-1)
| Entire | Name: Merozoite surface protein 1 (MSP-1) |
|---|---|
| Components |
|
-Supramolecule #1: Merozoite surface protein 1 (MSP-1)
| Supramolecule | Name: Merozoite surface protein 1 (MSP-1) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Heteromeric assembly of p83/30 fusion and p38/42 fusion of MSP-1 from Plasmodium falciparum (dimer). |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #1: Precursor of the major merozoite surface antigens
| Macromolecule | Name: Precursor of the major merozoite surface antigens / type: protein_or_peptide / ID: 1 Details: Fusion of the p83 and p30 subunits of the Merozoite surface protein-1 complex Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 101.973172 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VTHESYQELV KKLEALEDAV LTGYSLFQKE KMVLNEEEIT TKGASAQSGA SAQSGASAQS GASAQSGASA QSGASAQSGT SGPSGPSGT SPSSRSNTLP RSNTSSGASP PADASDSDAK SYADLKHRVR NYLFTIKELK YPELFDLTNH MLTLCDNIHG F KYLIDGYE ...String: VTHESYQELV KKLEALEDAV LTGYSLFQKE KMVLNEEEIT TKGASAQSGA SAQSGASAQS GASAQSGASA QSGASAQSGT SGPSGPSGT SPSSRSNTLP RSNTSSGASP PADASDSDAK SYADLKHRVR NYLFTIKELK YPELFDLTNH MLTLCDNIHG F KYLIDGYE EINELLYKLN FYFDLLRAKL NDVCANDYCQ IPFNLKIRAN ELDVLKKLVF GYRKPLDNIK DNVGKMEDYI KK NKTTIAN INELIEGSKK TIDQNKNADN EEGKKKLYQA QYDLSIYNKQ LEEAHNLISV LEKRIDTLKK NENIKKLLDK INE IKNPPP ANSGNTPNTL LDKNKKIEEH EEKIKEIAKT IKFNIDSLFT DPLELEYYLR EKNKKVDVTP KSQDPTKSVQ IPKV PYPNG IVYPLPLTDI HNSLAADNDK NSYGDLMNPH TKEKINEKII TDNKERKIFI NNIKKKIDLE EKNINHTKEQ NKKLL EDYE KSKKDYEELL EKFYEMKFNN NFDKDVVDKI FSARYTYNVE KQRYNNKFSS SNNSVYNVQK LKKALSYLED YSLRKG ISE KDFNHYYTLK TGLEADIKKL TEEIKSSENK ILEKNFKGLT HSANGSLEVS DIVKLQVQKV LLIKKIEDLR KIELFLK NA QLKDSIHVPN IYKPQNKPEP YYLIVLKKEV DKLKEFIPKV KDMLKKEQAV LSSITQPLVA ASETTEDGGH STHTLSQS G ETEVTEETEE TEETVGHTTT VTITLPPTQP SPPKEVKVVE NSIEQKSNDN SQALTKTVYL KKLDEFLTKS YICHKYILV SNSSMDQKLL EVYNLTPEEE NELKSCDPLD LLFNIQNNIP AMYSLYDSMN NDLQHLFFEL YQKEMIYYLH KLKEENHIKK LLEEQKQIT GT |
-Macromolecule #2: Merozoite surface protein-1
| Macromolecule | Name: Merozoite surface protein-1 / type: protein_or_peptide / ID: 2 Details: Fusion of the p38 and p42 subunits of the Merozoite surface protein-1 complex Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 89.615805 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SSTSSPGNTT VNTAQSATHS NSQNQQSNAS STNTQNGVAV SSGPAVVEES HDPLTVLSIS NDLKGIVSLL NLGNKTKVPN PLTISTTEM EKFYENILKN NDTYFNDDIK QFVKSNSKVI TGLTETQKNA LNDEIKKLKD TLQLSFDLYN KYKLKLDRLF N KKKELGQD ...String: SSTSSPGNTT VNTAQSATHS NSQNQQSNAS STNTQNGVAV SSGPAVVEES HDPLTVLSIS NDLKGIVSLL NLGNKTKVPN PLTISTTEM EKFYENILKN NDTYFNDDIK QFVKSNSKVI TGLTETQKNA LNDEIKKLKD TLQLSFDLYN KYKLKLDRLF N KKKELGQD KMQIKKLTLL KEQLESKLNS LNNPHNVLQN FSVFFNKKKE AEIAETENTL ENTKILLKHY KGLVKYYNGE SS PLKTLSE VSIQTEDNYA NLEKFRVLSK IDGKLNDNLH LGKKKLSFLS SGLHHLITEL KEVIKNKNYT GNSPSENNKK VNE ALKSYE NFLPEAKVTT VVTPPQPDVT PSPLSVRVSG SSGSTKEETQ IPTSGSLLTE LQQVVQLQNY DEEDDSLVVL PIFG ESEDN DEYLDQVVTG EAISVTMDNI LSGFENEYDV IYLKPLAGVY RSLKKQIEKN IFTFNLNLND ILNSRLKKRK YFLDV LESD LMQFKHISSN EYIIEDSFKL LNSEQKNTLL KSYKYIKESV ENDIKFAQEG ISYYEKVLAK YKDDLESIKK VIKEEK EKF PSSPPTTPPS PAKTDEQKKE SKFLPFLTNI ETLYNNLVNK IDDYLINLKA KINDCNVEKD EAHVKITKLS DLKAIDD KI DLFKNPYDFE AIKKLINDDT KKDMLGKLLS TGLVQNFPNT IISKLIEGKF QDMLNISQHQ CVKKQCPENS GCFRHLDE R EECKCLLNYK QEGDKCVENP NPTCNENNGG CDADATCTEE DSGSSRKKIT CECTKPDSYP LFDGIFCSSS N |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z
Z Y
Y X
X