+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10109 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of beta-Galactosidase from Thermotoga maritima | |||||||||
 Map data Map data | Map from Relion3 Refine3D after postprocessing using the deposited mask and automatic filtering/sharpening, and sharpened with LocScale | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | BETA-GALACTOSIDASE / CARBOHYDRATE / GLUCOSYL HYDROLASE / THERMOTOGA MARITIMA / TRANSGLYCOSYLATION / IMMOBILIZATION / CRYOEM / GRAPHENE-OXIDE / GALACTOOLIGOSACCHARIDES / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationlactose catabolic process / beta-galactosidase complex / beta-galactosidase / beta-galactosidase activity / carbohydrate binding Similarity search - Function | |||||||||
| Biological species |   Thermotoga maritima MSB8 (bacteria) Thermotoga maritima MSB8 (bacteria) | |||||||||
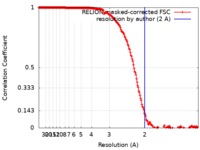
| Method | single particle reconstruction / cryo EM / Resolution: 2.0 Å | |||||||||
 Authors Authors | Miguez-Amil S / Jimenez-Ortega E | |||||||||
 Citation Citation |  Journal: ACS Chem Biol / Year: 2020 Journal: ACS Chem Biol / Year: 2020Title: The cryo-EM Structure of β-Galactosidase: Quaternary Structure Guides Protein Engineering. Authors: Samuel Míguez Amil / Elena Jiménez-Ortega / Mercedes Ramírez-Escudero / David Talens-Perales / Julia Marín-Navarro / Julio Polaina / Julia Sanz-Aparicio / Rafael Fernandez-Leiro /  Abstract: Lactose intolerance is a common digestive disorder that affects a large proportion of the adult human population. The severity of the symptoms is highly variable, depending on the susceptibility to ...Lactose intolerance is a common digestive disorder that affects a large proportion of the adult human population. The severity of the symptoms is highly variable, depending on the susceptibility to the sugar and the amount digested. For that reason, enzymes that can be used for the production of lactose-free milk and milk derivatives have acquired singular biotechnological importance. One such case is β-galactosidase (TmLac). Here, we report the cryo-EM structure of TmLac at 2.0 Å resolution. The protein features a newly solved domain at its C-terminus, characteristic of the genus , which promotes a peculiar octameric arrangement. We have assessed the constraints imposed by the quaternary protein structure on the construction of hybrid versions of this GH2 enzyme. Carbohydrate binding modules (CBM) from the CBM2 and CBM9 families have been added at either the amino or carboxy terminus, and the structural and functional effects of such modifications have been analyzed. The results provide a basis for the rational design of hybrid enzymes that can be efficiently attached to different solid supports. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10109.map.gz emd_10109.map.gz | 21.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10109-v30.xml emd-10109-v30.xml emd-10109.xml emd-10109.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10109_fsc.xml emd_10109_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_10109.png emd_10109.png | 166 KB | ||
| Masks |  emd_10109_msk_1.map emd_10109_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10109.cif.gz emd-10109.cif.gz | 6.9 KB | ||
| Others |  emd_10109_half_map_1.map.gz emd_10109_half_map_1.map.gz emd_10109_half_map_2.map.gz emd_10109_half_map_2.map.gz | 405.3 MB 405.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10109 http://ftp.pdbj.org/pub/emdb/structures/EMD-10109 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10109 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10109 | HTTPS FTP |
-Validation report
| Summary document |  emd_10109_validation.pdf.gz emd_10109_validation.pdf.gz | 916 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10109_full_validation.pdf.gz emd_10109_full_validation.pdf.gz | 915.6 KB | Display | |
| Data in XML |  emd_10109_validation.xml.gz emd_10109_validation.xml.gz | 27 KB | Display | |
| Data in CIF |  emd_10109_validation.cif.gz emd_10109_validation.cif.gz | 34.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10109 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10109 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10109 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10109 | HTTPS FTP |
-Related structure data
| Related structure data |  6s6zMC  6sd0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10109.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10109.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map from Relion3 Refine3D after postprocessing using the deposited mask and automatic filtering/sharpening, and sharpened with LocScale | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.67 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10109_msk_1.map emd_10109_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 from Relion3
| File | emd_10109_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 from Relion3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 from Relion3
| File | emd_10109_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 from Relion3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Map of beta-Galactosidase from Thermotoga maritima
| Entire | Name: Map of beta-Galactosidase from Thermotoga maritima |
|---|---|
| Components |
|
-Supramolecule #1: Map of beta-Galactosidase from Thermotoga maritima
| Supramolecule | Name: Map of beta-Galactosidase from Thermotoga maritima / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Map from Relion3: Refined 3D after postprocessing using the deposited mask and automatic filtering/sharpening. Sharpened by LocScale |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima MSB8 (bacteria) Thermotoga maritima MSB8 (bacteria) |
| Molecular weight | Theoretical: 1.02 MDa |
-Macromolecule #1: Beta-galactosidase
| Macromolecule | Name: Beta-galactosidase / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO / EC number: beta-galactosidase |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima MSB8 (bacteria) Thermotoga maritima MSB8 (bacteria) |
| Molecular weight | Theoretical: 127.653539 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PYEWENPQLV SEGTEKSHAS FIPYLDPFSG EWEYPEEFIS LNGNWRFLFA KNPFEVPEDF FSEKFDDSNW DEIEVPSNWE MKGYGKPIY TNVVYPFEPN PPFVPKDDNP TGVYRRWIEI PEDWFKKEIF LHFEGVRSFF YLWVNGKKIG FSKDSCTPAE F RLTDVLRP ...String: PYEWENPQLV SEGTEKSHAS FIPYLDPFSG EWEYPEEFIS LNGNWRFLFA KNPFEVPEDF FSEKFDDSNW DEIEVPSNWE MKGYGKPIY TNVVYPFEPN PPFVPKDDNP TGVYRRWIEI PEDWFKKEIF LHFEGVRSFF YLWVNGKKIG FSKDSCTPAE F RLTDVLRP GKNLITVEVL KWSDGSYLED QDMWWFAGIY RDVYLYALPK FHIRDVFVRT DLDENYRNGK IFLDVEMRNL GE EEEKDLE VTLITPDGDE KTLVKETVKP EDRVLSFAFD VKDPKKWSAE TPHLYVLKLK LGEDEKKVNF GFRKIEIKDG TLL FNGKPL YIKGVNRHEF DPDRGHAVTV ERMIQDIKLM KQHNINTVRT SHYPNQTKWY DLCDYFGLYV IDEANIESHG IDWD PEVTL ANRWEWEKAH FDRIKRMVER DKNHPSIIFW SLGNEAGDGV NFEKAALWIK KRDNTRLIHY EGTTRRGESY YVDVF SLMY PKMDILLEYA SKKREKPFIM CEYAHAMGNS VGNLKDYWDV IEKYPYLHGG CIWDWVDQGI RKKDENGREF WAYGGD FGD TPNDGNFCIN GVVLPDRTPE PELYEVKKVY QNVKIRQVSK DTYEVENRYL FTNLEMFDGA WKIRKDGEVI EEKTFKI FA EPGEKRLLKI PLPEMDDSEY FLEISFSLSE DTPWAEKGHV VAWEQFLLKA PAFEKKSISD GVSLREDGKH LTVEAKDT V YVFSKLTGLL EQILHRRKKI LKSPVVPNFW RVPTDNDIGN RMPQRLAIWK RASKERKLFK MHWKKEENRV SVHSVFQLP GNSWVYTTYT VFGNGDVLVD LSLIPAEDVP EIPRIGFQFT VPEEFGTVEW YGRGPHETYW DRKESGLFAR YRKAVGEMMH RYVRPQETG NRSDVRWFAL SDGETKLFVS GMPQIDFSVW PFSMEDLERV QHISELPERD FVTVNVDFRQ MGLGGDDSWG A MPHLEYRL LPKPYRFSFR MRISEEIPSW RVLAAIPETL HVEMSSEDVI REGDTLRVKF SLLNDTPLSK EKQVVLFVDG NE YSVRRVV IPPFKKEELV FKVEGLKKGE HLIHTNLNTR KTIYVR UniProtKB: Beta-galactosidase |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 8 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 1384 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Support film - Material: GRAPHENE OXIDE / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 30.0 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 3600 / Average exposure time: 25.0 sec. / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 2.6 µm / Calibrated defocus min: 0.4 µm / Calibrated magnification: 20895 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-6s6z: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)