[English] 日本語
 Yorodumi
Yorodumi- EMDB-0748: 298 K cryoEM structure of Sso-KARI in complex with Mg2+, NADH and CPD -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0748 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 298 K cryoEM structure of Sso-KARI in complex with Mg2+, NADH and CPD | |||||||||
 Map data Map data | Sso-KARI dodecameric enzyme in complex with Mg2 , NADH and CPD, and cryoEM sample was prepared at 298 K. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / ISOMERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationketol-acid reductoisomerase (NADP+) / ketol-acid reductoisomerase activity / L-valine biosynthetic process / isoleucine biosynthetic process / isomerase activity / metal ion binding Similarity search - Function | |||||||||
| Biological species |   Saccharolobus solfataricus (archaea) Saccharolobus solfataricus (archaea) | |||||||||
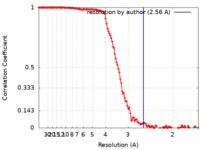
| Method | single particle reconstruction / cryo EM / Resolution: 2.56 Å | |||||||||
 Authors Authors | Chen CY / Chang YC / Lin BL / Huang CH / Tsai MD | |||||||||
 Citation Citation |  Journal: J Am Chem Soc / Year: 2019 Journal: J Am Chem Soc / Year: 2019Title: Temperature-Resolved Cryo-EM Uncovers Structural Bases of Temperature-Dependent Enzyme Functions. Authors: Chin-Yu Chen / Yuan-Chih Chang / Bo-Lin Lin / Chun-Hsiang Huang / Ming-Daw Tsai /  Abstract: Protein functions are temperature-dependent, but protein structures are usually solved at a single (often low) temperature because of limitations on the conditions of crystal growth or protein ...Protein functions are temperature-dependent, but protein structures are usually solved at a single (often low) temperature because of limitations on the conditions of crystal growth or protein vitrification. Here we demonstrate the feasibility of solving cryo-EM structures of proteins vitrified at high temperatures, solve 12 structures of an archaeal ketol-acid reductoisomerase (KARI) vitrified at 4-70 °C, and show that structures of both the Mg form (KARI:2Mg) and its ternary complex (KARI:2Mg:NADH:inhibitor) are temperature-dependent in correlation with the temperature dependence of enzyme activity. Furthermore, structural analyses led to dissection of the induced-fit mechanism into ligand-induced and temperature-induced effects and to capture of temperature-resolved intermediates of the temperature-induced conformational change. The results also suggest that it is preferable to solve cryo-EM structures of protein complexes at functional temperatures. These studies should greatly expand the landscapes of protein structure-function relationships and enhance the mechanistic analysis of enzymatic functions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0748.map.gz emd_0748.map.gz | 154 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0748-v30.xml emd-0748-v30.xml emd-0748.xml emd-0748.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0748_fsc.xml emd_0748_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_0748.png emd_0748.png | 226.4 KB | ||
| Filedesc metadata |  emd-0748.cif.gz emd-0748.cif.gz | 5.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0748 http://ftp.pdbj.org/pub/emdb/structures/EMD-0748 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0748 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0748 | HTTPS FTP |
-Validation report
| Summary document |  emd_0748_validation.pdf.gz emd_0748_validation.pdf.gz | 542.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0748_full_validation.pdf.gz emd_0748_full_validation.pdf.gz | 542.1 KB | Display | |
| Data in XML |  emd_0748_validation.xml.gz emd_0748_validation.xml.gz | 13.4 KB | Display | |
| Data in CIF |  emd_0748_validation.cif.gz emd_0748_validation.cif.gz | 17.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0748 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0748 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0748 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0748 | HTTPS FTP |
-Related structure data
| Related structure data |  6kpjMC  0740C  0742C  0743C  0746C  0747C  0749C  0750C  0751C  0752C  0753C  0754C  6kouC  6kpaC  6kpeC  6kphC  6kpiC  6kpkC  6kq4C  6kq8C  6kqjC  6kqkC  6kqoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0748.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0748.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sso-KARI dodecameric enzyme in complex with Mg2 , NADH and CPD, and cryoEM sample was prepared at 298 K. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : KARI-Mg2+/NADH/CPD complex
| Entire | Name: KARI-Mg2+/NADH/CPD complex |
|---|---|
| Components |
|
-Supramolecule #1: KARI-Mg2+/NADH/CPD complex
| Supramolecule | Name: KARI-Mg2+/NADH/CPD complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Saccharolobus solfataricus (archaea) Saccharolobus solfataricus (archaea) |
-Macromolecule #1: Ketol-acid reductoisomerase
| Macromolecule | Name: Ketol-acid reductoisomerase / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharolobus solfataricus (archaea) Saccharolobus solfataricus (archaea) |
| Molecular weight | Theoretical: 37.229855 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDKTVLDANL DPLKGKTIGV IGYGNQGRVQ ATIMRENGLN VIVGNVKDKY YELAKKEGFE VYEIDEAVRR SDVALLLIPD EVMKEVYEK KIAPVLQGKK EFVLDFASGY NVAFGLIRPP KSVDTIMVAP RMVGEGIMDL HKQGKGYPVL LGVKQDASGK A WDYAKAIA ...String: MDKTVLDANL DPLKGKTIGV IGYGNQGRVQ ATIMRENGLN VIVGNVKDKY YELAKKEGFE VYEIDEAVRR SDVALLLIPD EVMKEVYEK KIAPVLQGKK EFVLDFASGY NVAFGLIRPP KSVDTIMVAP RMVGEGIMDL HKQGKGYPVL LGVKQDASGK A WDYAKAIA KGIGAIPGGI AVISSFEEEA LLDLMSEHTW VPILFGAIKA CYDIAVKEYG VSPEAALLEF YASGELAEIA RL IAEEGIF NQMVHHSTTS QYGTLTRMFK YYDVVRRIVE NEAKYIWDGS FAKEWSLEQQ AGYPVFYRLW ELATQSEMAK AEK ELYKLL GRKVKND UniProtKB: Ketol-acid reductoisomerase |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 24 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE
| Macromolecule | Name: 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE / type: ligand / ID: 3 / Number of copies: 12 / Formula: NAI |
|---|---|
| Molecular weight | Theoretical: 665.441 Da |
| Chemical component information |  ChemComp-NAI: |
-Macromolecule #4: cyclopropane-1,1-dicarboxylic acid
| Macromolecule | Name: cyclopropane-1,1-dicarboxylic acid / type: ligand / ID: 4 / Number of copies: 12 / Formula: 9TY |
|---|---|
| Molecular weight | Theoretical: 130.099 Da |
| Chemical component information | 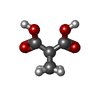 ChemComp-9TY: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)