+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDAL5 |
|---|---|
 Sample Sample | CD27L
|
| Biological species |   Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) |
 Citation Citation |  Journal: PLoS Pathog / Year: 2014 Journal: PLoS Pathog / Year: 2014Title: The CD27L and CTP1L endolysins targeting Clostridia contain a built-in trigger and release factor. Authors: Matthew Dunne / Haydyn D T Mertens / Vasiliki Garefalaki / Cy M Jeffries / Andrew Thompson / Edward A Lemke / Dmitri I Svergun / Melinda J Mayer / Arjan Narbad / Rob Meijers /    Abstract: The bacteriophage ΦCD27 is capable of lysing Clostridium difficile, a pathogenic bacterium that is a major cause for nosocomial infection. A recombinant CD27L endolysin lyses C. difficile in vitro, ...The bacteriophage ΦCD27 is capable of lysing Clostridium difficile, a pathogenic bacterium that is a major cause for nosocomial infection. A recombinant CD27L endolysin lyses C. difficile in vitro, and represents a promising alternative as a bactericide. To better understand the lysis mechanism, we have determined the crystal structure of an autoproteolytic fragment of the CD27L endolysin. The structure covers the C-terminal domain of the endolysin, and represents a novel fold that is identified in a number of lysins that target Clostridia bacteria. The structure indicates endolysin cleavage occurs at the stem of the linker connecting the catalytic domain with the C-terminal domain. We also solved the crystal structure of the C-terminal domain of a slow cleaving mutant of the CTP1L endolysin that targets C. tyrobutyricum. Two distinct dimerization modes are observed in the crystal structures for both endolysins, despite a sequence identity of only 22% between the domains. The dimers are validated to be present for the full length protein in solution by right angle light scattering, small angle X-ray scattering and cross-linking experiments using the cross-linking amino acid p-benzoyl-L-phenylalanine (pBpa). Mutagenesis on residues contributing to the dimer interfaces indicates that there is a link between the dimerization modes and the autocleavage mechanism. We show that for the CTP1L endolysin, there is a reduction in lysis efficiency that is proportional to the cleavage efficiency. We propose a model for endolysin triggering, where the extended dimer presents the inactive state, and a switch to the side-by-side dimer triggers the cleavage of the C-terminal domain. This leads to the release of the catalytic portion of the endolysin, enabling the efficient digestion of the bacterial cell wall. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
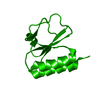
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Models
| Model #269 |  Type: atomic / Radius of dummy atoms: 1.90 A / Comment: OLIGOMER component 1: head-head CD27L dimer / Chi-square value: 2.4649  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #270 |  Type: atomic / Radius of dummy atoms: 1.90 A / Comment: OLIGOMER component 2: side-side CD27L dimer / Chi-square value: 2.4649  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #271 |  Type: atomic / Radius of dummy atoms: 1.90 A / Comment: OLIGOMER component 3a / Chi-square value: 2.4649  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #272 |  Type: atomic / Radius of dummy atoms: 1.90 A / Comment: OLIGOMER component 3b / Chi-square value: 2.4649  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #273 |  Type: atomic / Radius of dummy atoms: 1.90 A / Chi-square value: 2.4649  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #274 |  Type: atomic / Radius of dummy atoms: 1.90 A / Comment: OLIGOMER component 3c / Chi-square value: 2.4649  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #275 |  Type: atomic / Radius of dummy atoms: 1.90 A / Comment: OLIGOMER component 3d / Chi-square value: 2.4649  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #276 |  Type: atomic / Radius of dummy atoms: 1.90 A / Comment: OLIGOMER component 3e / Chi-square value: 2.4649  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: CD27L / Sample MW: 64.2 kDa / Specimen concentration: 0.90-4.00 / Concentration method: A280 |
|---|---|
| Buffer | Name: 20 mM HEPES 150 mM NaCl / pH: 7.5 / Composition: 150 mM NaCl |
| Entity #94 | Name: CD27L / Type: protein Description: Clostridium difficile bacteriophage 27 endolysin Formula weight: 32.1 / Num. of mol.: 2 / Source: Clostridioides difficile Sequence: MGSSHHHHHH SSGLVPRGSH MKICITVGHS ILKSGACTSA DGVVNEYQYN KSLAPVLADT FRKEGHKVDV IIsPEKQFKT KNEEKSYKIP RVNSGGYDLL IELHLNASNG QGKGSEVLYY SNKGLEYATR ICDKLGTVFK NRGAKLDKRL YILNSSKPTA VLIESFFCDN ...Sequence: MGSSHHHHHH SSGLVPRGSH MKICITVGHS ILKSGACTSA DGVVNEYQYN KSLAPVLADT FRKEGHKVDV IIsPEKQFKT KNEEKSYKIP RVNSGGYDLL IELHLNASNG QGKGSEVLYY SNKGLEYATR ICDKLGTVFK NRGAKLDKRL YILNSSKPTA VLIESFFCDN KEDYDKAKKL GHEGIAKLIV EGVLNKNINN EGVKQMYKHT IVYDGEVDKI SATVVGWGYN DGKILICDIK DYVPGQTQNL YVVGGGACEK ISSITKEKFI MIKGNDRFDT LYKALDFIN |
-Experimental information
| Beam | Instrument name:  DORIS III X33 DORIS III X33  / City: Hamburg / 国: Germany / City: Hamburg / 国: Germany  / Shape / Shape : 0.6 / Type of source: X-ray synchrotron : 0.6 / Type of source: X-ray synchrotron Synchrotron / Wavelength: 0.15 Å / Dist. spec. to detc.: 2.7 mm Synchrotron / Wavelength: 0.15 Å / Dist. spec. to detc.: 2.7 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M-W / Pixsize x: 0.172 mm | ||||||||||||||||||||||||||||||
| Scan |
| ||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller


 SASDAL5
SASDAL5