+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8sl4 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
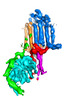
| Title | Dimeric form of human adenylyl cyclase 5 | ||||||||||||||||||
 Components Components | Adenylate cyclase type 5 | ||||||||||||||||||
 Keywords Keywords |  SIGNALING PROTEIN / SIGNALING PROTEIN /  Adenylyl cyclase Adenylyl cyclase | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationAdenylate cyclase activating pathway / adenylate cyclase-inhibiting dopamine receptor signaling pathway / G protein-coupled adenosine receptor signaling pathway /  adenylate cyclase / regulation of insulin secretion involved in cellular response to glucose stimulus / cAMP biosynthetic process / adenylate cyclase / regulation of insulin secretion involved in cellular response to glucose stimulus / cAMP biosynthetic process /  adenylate cyclase activity / PKA activation / PKA activation in glucagon signalling / neuromuscular process controlling balance ...Adenylate cyclase activating pathway / adenylate cyclase-inhibiting dopamine receptor signaling pathway / G protein-coupled adenosine receptor signaling pathway / adenylate cyclase activity / PKA activation / PKA activation in glucagon signalling / neuromuscular process controlling balance ...Adenylate cyclase activating pathway / adenylate cyclase-inhibiting dopamine receptor signaling pathway / G protein-coupled adenosine receptor signaling pathway /  adenylate cyclase / regulation of insulin secretion involved in cellular response to glucose stimulus / cAMP biosynthetic process / adenylate cyclase / regulation of insulin secretion involved in cellular response to glucose stimulus / cAMP biosynthetic process /  adenylate cyclase activity / PKA activation / PKA activation in glucagon signalling / neuromuscular process controlling balance / adenylate cyclase activity / PKA activation / PKA activation in glucagon signalling / neuromuscular process controlling balance /  adenylate cyclase binding / Adenylate cyclase inhibitory pathway / Hedgehog 'off' state / cellular response to forskolin / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / FCGR3A-mediated IL10 synthesis / locomotory behavior / adenylate cyclase binding / Adenylate cyclase inhibitory pathway / Hedgehog 'off' state / cellular response to forskolin / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / FCGR3A-mediated IL10 synthesis / locomotory behavior /  cilium / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Glucagon signaling in metabolic regulation / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / G alpha (i) signalling events / positive regulation of cytosolic calcium ion concentration / G alpha (s) signalling events / cilium / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Glucagon signaling in metabolic regulation / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / G alpha (i) signalling events / positive regulation of cytosolic calcium ion concentration / G alpha (s) signalling events /  scaffold protein binding / intracellular signal transduction / scaffold protein binding / intracellular signal transduction /  ATP binding / ATP binding /  membrane / membrane /  metal ion binding / metal ion binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 7 Å cryo EM / Resolution: 7 Å | ||||||||||||||||||
 Authors Authors | Yen, Y.C. / Tesmer, J.J.G. | ||||||||||||||||||
| Funding support |  United States, 5items United States, 5items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structure of adenylyl cyclase 5 in complex with Gβγ offers insights into ADCY5-related dyskinesia. Authors: Yu-Chen Yen / Yong Li / Chun-Liang Chen / Thomas Klose / Val J Watts / Carmen W Dessauer / John J G Tesmer /  Abstract: The nine different membrane-anchored adenylyl cyclase isoforms (AC1-9) in mammals are stimulated by the heterotrimeric G protein, Gα, but their response to Gβγ regulation is isoform specific. In ...The nine different membrane-anchored adenylyl cyclase isoforms (AC1-9) in mammals are stimulated by the heterotrimeric G protein, Gα, but their response to Gβγ regulation is isoform specific. In the present study, we report cryo-electron microscope structures of ligand-free AC5 in complex with Gβγ and a dimeric form of AC5 that could be involved in its regulation. Gβγ binds to a coiled-coil domain that links the AC transmembrane region to its catalytic core as well as to a region (C) that is known to be a hub for isoform-specific regulation. We confirmed the Gβγ interaction with both purified proteins and cell-based assays. Gain-of-function mutations in AC5 associated with human familial dyskinesia are located at the interface of AC5 with Gβγ and show reduced conditional activation by Gβγ, emphasizing the importance of the observed interaction for motor function in humans. We propose a molecular mechanism wherein Gβγ either prevents dimerization of AC5 or allosterically modulates the coiled-coil domain, and hence the catalytic core. As our mechanistic understanding of how individual AC isoforms are uniquely regulated is limited, studies such as this may provide new avenues for isoform-specific drug development. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8sl4.cif.gz 8sl4.cif.gz | 641.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8sl4.ent.gz pdb8sl4.ent.gz | 528.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8sl4.json.gz 8sl4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/sl/8sl4 https://data.pdbj.org/pub/pdb/validation_reports/sl/8sl4 ftp://data.pdbj.org/pub/pdb/validation_reports/sl/8sl4 ftp://data.pdbj.org/pub/pdb/validation_reports/sl/8sl4 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  40573MC  8sl3C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 139066.312 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: ADCY5 / Production host: Homo sapiens (human) / Gene: ADCY5 / Production host:  Mammalia (mammals) / References: UniProt: O95622, Mammalia (mammals) / References: UniProt: O95622,  adenylate cyclase adenylate cyclase |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Dimeric form of human adenylyl cyclase 5 / Type: COMPLEX / Entity ID: all / Source: RECOMBINANT | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.28 MDa / Experimental value: NO | ||||||||||||||||||||||||||||||
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||||||||
| Source (recombinant) | Organism:  Mammalia (mammals) Mammalia (mammals) | ||||||||||||||||||||||||||||||
| Buffer solution | pH: 7.4 | ||||||||||||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||||||||||||
| Specimen | Conc.: 0.2 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES | ||||||||||||||||||||||||||||||
| Specimen support | Grid type: Quantifoil R1.2/1.3 | ||||||||||||||||||||||||||||||
Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3000 nm / Nominal defocus min: 1000 nm / Cs Bright-field microscopy / Nominal defocus max: 3000 nm / Nominal defocus min: 1000 nm / Cs : 2.7 mm / C2 aperture diameter: 100 µm : 2.7 mm / C2 aperture diameter: 100 µm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 53.8 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| EM software |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||
3D reconstruction | Resolution: 7 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 102581 / Symmetry type: POINT |
 Movie
Movie Controller
Controller



 PDBj
PDBj
















