+ Open data
Open data
- Basic information
Basic information
| Entry | 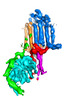 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
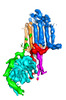
| Title | Human adenylyl Cyclase 5 in complex with Gbg | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords |  Adenylyl cyclase / Adenylyl cyclase /  G-proteins / G-proteins /  complex / complex /  SIGNALING PROTEIN SIGNALING PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationAdenylate cyclase activating pathway / adenylate cyclase-inhibiting dopamine receptor signaling pathway / G protein-coupled adenosine receptor signaling pathway /  adenylate cyclase / regulation of insulin secretion involved in cellular response to glucose stimulus / cAMP biosynthetic process / adenylate cyclase / regulation of insulin secretion involved in cellular response to glucose stimulus / cAMP biosynthetic process /  adenylate cyclase activity / PKA activation / PKA activation in glucagon signalling / neuromuscular process controlling balance ...Adenylate cyclase activating pathway / adenylate cyclase-inhibiting dopamine receptor signaling pathway / G protein-coupled adenosine receptor signaling pathway / adenylate cyclase activity / PKA activation / PKA activation in glucagon signalling / neuromuscular process controlling balance ...Adenylate cyclase activating pathway / adenylate cyclase-inhibiting dopamine receptor signaling pathway / G protein-coupled adenosine receptor signaling pathway /  adenylate cyclase / regulation of insulin secretion involved in cellular response to glucose stimulus / cAMP biosynthetic process / adenylate cyclase / regulation of insulin secretion involved in cellular response to glucose stimulus / cAMP biosynthetic process /  adenylate cyclase activity / PKA activation / PKA activation in glucagon signalling / neuromuscular process controlling balance / adenylate cyclase activity / PKA activation / PKA activation in glucagon signalling / neuromuscular process controlling balance /  adenylate cyclase binding / Adenylate cyclase inhibitory pathway / Hedgehog 'off' state / cellular response to forskolin / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / FCGR3A-mediated IL10 synthesis / locomotory behavior / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / adenylate cyclase binding / Adenylate cyclase inhibitory pathway / Hedgehog 'off' state / cellular response to forskolin / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / FCGR3A-mediated IL10 synthesis / locomotory behavior / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits /  cilium / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / adenylate cyclase-activating G protein-coupled receptor signaling pathway / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / cilium / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / adenylate cyclase-activating G protein-coupled receptor signaling pathway / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding /  heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade /  extracellular vesicle / G alpha (12/13) signalling events / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / extracellular vesicle / G alpha (12/13) signalling events / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye /  GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / positive regulation of cytosolic calcium ion concentration / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / positive regulation of cytosolic calcium ion concentration / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events /  scaffold protein binding / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / intracellular signal transduction / G protein-coupled receptor signaling pathway / lysosomal membrane / scaffold protein binding / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / intracellular signal transduction / G protein-coupled receptor signaling pathway / lysosomal membrane /  GTPase activity / GTPase activity /  synapse / protein-containing complex binding / synapse / protein-containing complex binding /  signal transduction / extracellular exosome / signal transduction / extracellular exosome /  ATP binding / ATP binding /  membrane / membrane /  metal ion binding / metal ion binding /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 7.0 Å cryo EM / Resolution: 7.0 Å | ||||||||||||||||||
 Authors Authors | Yen YC / Tesmer JJG | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structure of adenylyl cyclase 5 in complex with Gβγ offers insights into ADCY5-related dyskinesia. Authors: Yu-Chen Yen / Yong Li / Chun-Liang Chen / Thomas Klose / Val J Watts / Carmen W Dessauer / John J G Tesmer /  Abstract: The nine different membrane-anchored adenylyl cyclase isoforms (AC1-9) in mammals are stimulated by the heterotrimeric G protein, Gα, but their response to Gβγ regulation is isoform specific. In ...The nine different membrane-anchored adenylyl cyclase isoforms (AC1-9) in mammals are stimulated by the heterotrimeric G protein, Gα, but their response to Gβγ regulation is isoform specific. In the present study, we report cryo-electron microscope structures of ligand-free AC5 in complex with Gβγ and a dimeric form of AC5 that could be involved in its regulation. Gβγ binds to a coiled-coil domain that links the AC transmembrane region to its catalytic core as well as to a region (C) that is known to be a hub for isoform-specific regulation. We confirmed the Gβγ interaction with both purified proteins and cell-based assays. Gain-of-function mutations in AC5 associated with human familial dyskinesia are located at the interface of AC5 with Gβγ and show reduced conditional activation by Gβγ, emphasizing the importance of the observed interaction for motor function in humans. We propose a molecular mechanism wherein Gβγ either prevents dimerization of AC5 or allosterically modulates the coiled-coil domain, and hence the catalytic core. As our mechanistic understanding of how individual AC isoforms are uniquely regulated is limited, studies such as this may provide new avenues for isoform-specific drug development. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40572.map.gz emd_40572.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40572-v30.xml emd-40572-v30.xml emd-40572.xml emd-40572.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40572.png emd_40572.png | 72.8 KB | ||
| Filedesc metadata |  emd-40572.cif.gz emd-40572.cif.gz | 6.9 KB | ||
| Others |  emd_40572_half_map_1.map.gz emd_40572_half_map_1.map.gz emd_40572_half_map_2.map.gz emd_40572_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40572 http://ftp.pdbj.org/pub/emdb/structures/EMD-40572 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40572 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40572 | HTTPS FTP |
-Related structure data
| Related structure data |  8sl3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40572.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40572.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_40572_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_40572_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human adenylyl cyclase 5 in complex with Gbg
| Entire | Name: Human adenylyl cyclase 5 in complex with Gbg |
|---|---|
| Components |
|
-Supramolecule #1: Human adenylyl cyclase 5 in complex with Gbg
| Supramolecule | Name: Human adenylyl cyclase 5 in complex with Gbg / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 180 KDa |
-Macromolecule #1: Adenylate cyclase type 5
| Macromolecule | Name: Adenylate cyclase type 5 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number:  adenylate cyclase adenylate cyclase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 139.066312 KDa |
| Recombinant expression | Organism:  Mammalia (mammals) Mammalia (mammals) |
| Sequence | String: MSGSKSVSPP GYAAQKTAAP APRGGPEHRS AWGEADSRAN GYPHAPGGSA RGSTKKPGGA VTPQQQQRLA SRWRSDDDDD PPLSGDDPL AGGFGFSFRS KSAWQERGGD DCGRGSRRQR RGAASGGSTR APPAGGGGGS AAAAASAGGT EVRPRSVEVG L EERRGKGR ...String: MSGSKSVSPP GYAAQKTAAP APRGGPEHRS AWGEADSRAN GYPHAPGGSA RGSTKKPGGA VTPQQQQRLA SRWRSDDDDD PPLSGDDPL AGGFGFSFRS KSAWQERGGD DCGRGSRRQR RGAASGGSTR APPAGGGGGS AAAAASAGGT EVRPRSVEVG L EERRGKGR AADELEAGAV EGGEGSGDGG SSADSGSGAG PGAVLSLGAC CLALLQIFRS KKFPSDKLER LYQRYFFRLN QS SLTMLMA VLVLVCLVML AFHAARPPLQ LPYLAVLAAA VGVILIMAVL CNRAAFHQDH MGLACYALIA VVLAVQVVGL LLP QPRSAS EGIWWTVFFI YTIYTLLPVR MRAAVLSGVL LSALHLAIAL RTNAQDQFLL KQLVSNVLIF SCTNIVGVCT HYPA EVSQR QAFQETRECI QARLHSQREN QQQERLLLSV LPRHVAMEMK ADINAKQEDM MFHKIYIQKH DNVSILFADI EGFTS LASQ CTAQELVMTL NELFARFDKL AAENHCLRIK ILGDCYYCVS GLPEARADHA HCCVEMGMDM IEAISLVREV TGVNVN MRV GIHSGRVHCG VLGLRKWQFD VWSNDVTLAN HMEAGGKAGR IHITKATLNY LNGDYEVEPG CGGERNAYLK EHSIETF LI LRCTQKRKEE KAMIAKMNRQ RTNSIGHNPP HWGAERPFYN HLGGNQVSKE MKRMGFEDPK DKNAQESANP EDEVDEFL G RAIDARSIDR LRSEHVRKFL LTFREPDLEK KYSKQVDDRF GAYVACASLV FLFICFVQIT IVPHSIFMLS FYLTCSLLL TLVVFVSVIY SCVKLFPSPL QTLSRKIVRS KMNSTLVGVF TITLVFLAAF VNMFTCNSRD LLGCLAQEHN ISASQVNACH VAESAVNYS LGDEQGFCGS PWPNCNFPEY FTYSVLLSLL ACSVFLQISC IGKLVLMLAI ELIYVLIVEV PGVTLFDNAD L LVTANAID FFNNGTSQCP EHATKVALKV VTPIIISVFV LALYLHAQQV ESTARLDFLW KLQATEEKEE MEELQAYNRR LL HNILPKD VAAHFLARER RNDELYYQSC ECVAVMFASI ANFSEFYVEL EANNEGVECL RLLNEIIADF DEIISEDRFR QLE KIKTIG STYMAASGLN DSTYDKVGKT HIKALADFAM KLMDQMKYIN EHSFNNFQMK IGLNIGPVVA GVIGARKPQY DIWG NTVNV ASRMDSTGVP DRIQVTTDMY QVLAANTYQL ECRGVVKVKG KGEMMTYFLN GGPPLS UniProtKB: Adenylate cyclase type 5 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.41693 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: GERAN-8-YL GERAN
| Macromolecule | Name: GERAN-8-YL GERAN / type: ligand / ID: 4 / Number of copies: 1 / Formula: GER |
|---|---|
| Molecular weight | Theoretical: 274.484 Da |
| Chemical component information | 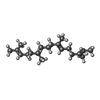 ChemComp-GER: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 53.8 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 1251384 |
|---|---|
| Startup model | Type of model: NONE |
| Initial angle assignment | Type: COMMON LINE / Software - Name: cryoSPARC |
| Final angle assignment | Type: COMMON LINE / Software - Name: cryoSPARC |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 7.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 61870 |
 Movie
Movie Controller
Controller
























 Z
Z Y
Y X
X