[English] 日本語
 Yorodumi
Yorodumi- PDB-8gcp: Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8gcp | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  GPCR complex GPCR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of eosinophil extravasation /  prostaglandin E receptor activity / Prostanoid ligand receptors / negative regulation of integrin activation / response to nematode / T-helper cell differentiation / prostaglandin E receptor activity / Prostanoid ligand receptors / negative regulation of integrin activation / response to nematode / T-helper cell differentiation /  regulation of stress fiber assembly / negative regulation of cytokine production / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex ...negative regulation of eosinophil extravasation / regulation of stress fiber assembly / negative regulation of cytokine production / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex ...negative regulation of eosinophil extravasation /  prostaglandin E receptor activity / Prostanoid ligand receptors / negative regulation of integrin activation / response to nematode / T-helper cell differentiation / prostaglandin E receptor activity / Prostanoid ligand receptors / negative regulation of integrin activation / response to nematode / T-helper cell differentiation /  regulation of stress fiber assembly / negative regulation of cytokine production / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex / regulation of stress fiber assembly / negative regulation of cytokine production / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex /  regulation of ossification / regulation of cAMP-mediated signaling / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / response to mechanical stimulus / regulation of mitotic spindle organization / cellular response to forskolin / JNK cascade / ERK1 and ERK2 cascade / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / Regulation of insulin secretion / G protein-coupled receptor binding / positive regulation of cytokine production / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / regulation of ossification / regulation of cAMP-mediated signaling / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / response to mechanical stimulus / regulation of mitotic spindle organization / cellular response to forskolin / JNK cascade / ERK1 and ERK2 cascade / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / Regulation of insulin secretion / G protein-coupled receptor binding / positive regulation of cytokine production / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / adenylate cyclase-modulating G protein-coupled receptor signaling pathway /  bone development / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / adenylate cyclase-activating G protein-coupled receptor signaling pathway / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Sensory perception of sweet, bitter, and umami (glutamate) taste / response to peptide hormone / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / negative regulation of inflammatory response / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / positive regulation of inflammatory response / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to mechanical stimulus / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / GDP binding / G-protein beta-subunit binding / bone development / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / adenylate cyclase-activating G protein-coupled receptor signaling pathway / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Sensory perception of sweet, bitter, and umami (glutamate) taste / response to peptide hormone / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / negative regulation of inflammatory response / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / positive regulation of inflammatory response / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to mechanical stimulus / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / GDP binding / G-protein beta-subunit binding /  heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade /  extracellular vesicle / G alpha (12/13) signalling events / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / extracellular vesicle / G alpha (12/13) signalling events / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye /  GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway /  cell cortex / midbody / positive regulation of cytosolic calcium ion concentration / G alpha (i) signalling events / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / response to lipopolysaccharide / Extra-nuclear estrogen signaling / cell cortex / midbody / positive regulation of cytosolic calcium ion concentration / G alpha (i) signalling events / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / response to lipopolysaccharide / Extra-nuclear estrogen signaling /  immune response / immune response /  cell cycle / cell cycle /  inflammatory response / G protein-coupled receptor signaling pathway / inflammatory response / G protein-coupled receptor signaling pathway /  cell division / lysosomal membrane / cell division / lysosomal membrane /  GTPase activity / GTPase activity /  centrosome / centrosome /  synapse / protein-containing complex binding / GTP binding / synapse / protein-containing complex binding / GTP binding /  nucleolus / magnesium ion binding / nucleolus / magnesium ion binding /  signal transduction / extracellular exosome / signal transduction / extracellular exosome /  nucleoplasm nucleoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.1 Å cryo EM / Resolution: 3.1 Å | ||||||
 Authors Authors | Huang, S.M. / Xiong, M.Y. / Liu, L. / Mu, J. / Sheng, C. / Sun, J. | ||||||
| Funding support |  China, 1items China, 1items
| ||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Single hormone or synthetic agonist induces G/G coupling selectivity of EP receptors via distinct binding modes and propagating paths. Authors: Shen-Ming Huang / Meng-Yao Xiong / Lei Liu / Jianqiang Mu / Ming-Wei Wang / Ying-Li Jia / Kui Cai / Lu Tie / Chao Zhang / Sheng Cao / Xin Wen / Jia-Le Wang / Sheng-Chao Guo / Yu Li / Chang- ...Authors: Shen-Ming Huang / Meng-Yao Xiong / Lei Liu / Jianqiang Mu / Ming-Wei Wang / Ying-Li Jia / Kui Cai / Lu Tie / Chao Zhang / Sheng Cao / Xin Wen / Jia-Le Wang / Sheng-Chao Guo / Yu Li / Chang-Xiu Qu / Qing-Tao He / Bo-Yang Cai / Chenyang Xue / Shiyi Gan / Yihe Xie / Xin Cong / Zhao Yang / Wei Kong / Shuo Li / Zijian Li / Peng Xiao / Fan Yang / Xiao Yu / You-Fei Guan / Xiaoyan Zhang / Zhongmin Liu / Bao-Xue Yang / Yang Du / Jin-Peng Sun /  Abstract: To accomplish concerted physiological reactions, nature has diversified functions of a single hormone at at least two primary levels: 1) Different receptors recognize the same hormone, and 2) ...To accomplish concerted physiological reactions, nature has diversified functions of a single hormone at at least two primary levels: 1) Different receptors recognize the same hormone, and 2) different cellular effectors couple to the same hormone-receptor pair [R.P. Xiao, , re15 (2001); L. Hein, J. D. Altman, B.K. Kobilka, , 181-184 (1999); Y. Daaka, L. M. Luttrell, R. J. Lefkowitz, , 88-91 (1997)]. Not only these questions lie in the heart of hormone actions and receptor signaling but also dissecting mechanisms underlying these questions could offer therapeutic routes for refractory diseases, such as kidney injury (KI) or X-linked nephrogenic diabetes insipidus (NDI). Here, we identified that G-biased signaling, but not G activation downstream of EP4, showed beneficial effects for both KI and NDI treatments. Notably, by solving Cryo-electron microscope (cryo-EM) structures of EP3-G, EP4-G, and EP4-G in complex with endogenous prostaglandin E (PGE)or two synthetic agonists and comparing with PGE-EP2-G structures, we found that unique primary sequences of prostaglandin E2 receptor (EP) receptors and distinct conformational states of the EP4 ligand pocket govern the G/G transducer coupling selectivity through different structural propagation paths, especially via TM6 and TM7, to generate selective cytoplasmic structural features. In particular, the orientation of the PGE ω-chain and two distinct pockets encompassing agonist L902688 of EP4 were differentiated by their G/G coupling ability. Further, we identified common and distinct features of cytoplasmic side of EP receptors for G/G coupling and provide a structural basis for selective and biased agonist design of EP4 with therapeutic potential. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8gcp.cif.gz 8gcp.cif.gz | 208.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8gcp.ent.gz pdb8gcp.ent.gz | 152.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8gcp.json.gz 8gcp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gc/8gcp https://data.pdbj.org/pub/pdb/validation_reports/gc/8gcp ftp://data.pdbj.org/pub/pdb/validation_reports/gc/8gcp ftp://data.pdbj.org/pub/pdb/validation_reports/gc/8gcp | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  29940MC  8gcmC  8gd9C  8gdaC  8gdbC  8gdcC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Guanine nucleotide-binding protein ... , 3 types, 3 molecules ABC
| #1: Protein | Mass: 40445.059 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: GNAI1 / Production host: Homo sapiens (human) / Gene: GNAI1 / Production host:   Baculovirus expression vector pFastBac1-HM / References: UniProt: P63096 Baculovirus expression vector pFastBac1-HM / References: UniProt: P63096 |
|---|---|
| #2: Protein | Mass: 39418.086 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: GNB1 / Production host: Homo sapiens (human) / Gene: GNB1 / Production host:   Baculovirus expression vector pFastBac1-HM / References: UniProt: P62873 Baculovirus expression vector pFastBac1-HM / References: UniProt: P62873 |
| #3: Protein | Mass: 12104.898 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: mRhiFer1_005810 / Production host: Homo sapiens (human) / Gene: mRhiFer1_005810 / Production host:   Baculovirus expression vector pFastBac1-HM / References: UniProt: A0A7J7XNR4 Baculovirus expression vector pFastBac1-HM / References: UniProt: A0A7J7XNR4 |
-Antibody / Protein / Non-polymers , 3 types, 3 molecules ER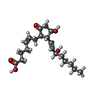

| #4: Antibody |  Single-chain variable fragment Single-chain variable fragmentMass: 32898.781 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Production host: Homo sapiens (human) / Production host:   Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
|---|---|
| #5: Protein | Mass: 53173.336 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: PTGER4, PTGER2 / Production host: Homo sapiens (human) / Gene: PTGER4, PTGER2 / Production host:   Baculovirus expression vector pFastBac1-HM / References: UniProt: P35408 Baculovirus expression vector pFastBac1-HM / References: UniProt: P35408 |
| #6: Chemical | ChemComp-P2E / ( Prostaglandin E2 Prostaglandin E2 |
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Cryo-EM Structure of Prostaglandin E Receptor EP4 Coupled to G Protein Type: COMPLEX / Entity ID: #1-#5 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:   Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Buffer solution | pH: 7.4 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : OTHER / Accelerating voltage: 300 kV / Illumination mode: OTHER : OTHER / Accelerating voltage: 300 kV / Illumination mode: OTHER |
| Electron lens | Mode: OTHER / Nominal defocus max: 2000 nm / Nominal defocus min: 800 nm |
| Image recording | Electron dose: 25 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
3D reconstruction | Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 291813 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller







 PDBj
PDBj






























